Here, Venkatesh et al V体育平台登录. investigated the p53-independent roles of MDMX and the MDM2–MDMX complex. They found that MDM2 and MDMX facilitate ferroptosis in cells with or without p53, and that PPARα activity is essential for MDM2 and MDMX to promote ferroptosis, suggesting that the MDM2–MDMX complex regulates lipids through altering PPARα activity.
Keywords: ferroptosis, MDM2, MDMX, p53-independent, PPARα, lipid metabolism, FSP1, CoQ10, cancer
Abstract
MDM2 and MDMX, negative regulators of the tumor suppressor p53, can work separately and as a heteromeric complex to restrain p53's functions. MDM2 also has pro-oncogenic roles in cells, tissues, and animals that are independent of p53. There is less information available about p53-independent roles of MDMX or the MDM2–MDMX complex. We found that MDM2 and MDMX facilitate ferroptosis in cells with or without p53 V体育官网入口. Using small molecules, RNA interference reagents, and mutant forms of MDMX, we found that MDM2 and MDMX, likely working in part as a complex, normally facilitate ferroptotic death. We observed that MDM2 and MDMX alter the lipid profile of cells to favor ferroptosis. Inhibition of MDM2 or MDMX leads to increased levels of FSP1 protein and a consequent increase in the levels of coenzyme Q10, an endogenous lipophilic antioxidant. This suggests that MDM2 and MDMX normally prevent cells from mounting an adequate defense against lipid peroxidation and thereby promote ferroptosis. Moreover, we found that PPARα activity is essential for MDM2 and MDMX to promote ferroptosis, suggesting that the MDM2–MDMX complex regulates lipids through altering PPARα activity. These findings reveal the complexity of cellular responses to MDM2 and MDMX and suggest that MDM2–MDMX inhibition might be useful for preventing degenerative diseases involving ferroptosis. Furthermore, they suggest that MDM2/MDMX amplification may predict sensitivity of some cancers to ferroptosis inducers.
Ferroptosis, an iron-dependent form of nonapoptotic cell death, occurs through an increase in cellular phospholipid peroxidation in the context of a compromised phospholipid peroxide repair system. Four main classes of ferroptosis inducers (FINs) are able to cause cell death through distinct mechanisms. The end result of the cellular responses to each FIN is ultimately cellular lipid peroxidation leading to cell death. Class I FINs (e. g. , erastin, its more potent derivative IKE (Larraufie et al. 2015), as well as sulfasalazine and sorafenib) inhibit system Xc− activity and deplete glutathione. Class II FINs [e. g. , (1S,3R)-RSL3, hereafter referred to as RSL3] directly inhibit the activity of GPX4, a glutathione-dependent phospholipid peroxidase. Class III FINs (e. g VSports在线直播. , FIN56) induce degradation of GPX4 and also interfere with the production of mevalonate pathway intermediates, including coenzyme Q10 (CoQ10). Finally, class IV FINs (e. g. , FINO2) stimulate lipid peroxidation, as well as the loss of GPX4 activity (Stockwell et al. 2017).
TP53 has been shown to increase sensitivity to ferroptosis through regulation of a number of downstream targets (Murphy 2016). First, p53 can decrease the expression of SLC7A11, a component of system Xc− that is necessary for uptake of cystine by some cells to generate intracellular cysteine; cysteine is in turn needed for the generation of glutathione, which is a substrate for the activity of GPX4 needed to suppress ferroptosis. Second, induction of GLS2, a p53 target gene that favors glutaminolysis, promotes ferroptosis V体育2025版. Third, p53 can induce SAT1, which favors the formation of lipid peroxides to drive this type of cell death.
However, other studies have provided evidence that p53 can suppress ferroptosis in some contexts. For example, p21, a well-studied p53 target, can alter cellular metabolism to retard ferroptosis (Tarangelo et al. 2018). Furthermore, through its protein–protein interaction with DPP4 that controls membrane lipid peroxidation (Xie et al. 2017), p53 can inhibit ferroptosis. Thus, the role of p53 in regulating ferroptosis can vary with context. Oncogenic alterations in the p53 network may modulate ferroptosis sensitivity through diverse mechanisms, contributing to either tumor suppression or oncogenesis VSports. These varying effects of p53 on ferroptosis led us to examine whether other components of the p53 network could also regulate ferroptosis.
Of key importance in the p53 network are the two negative regulators of p53, the homologous proteins MDM2 and MDMX. MDM2 is an E3 ligase that can ubiquitinate p53, as well as MDMX. While MDMX does not have E3 ligase activity on its own, it can form a heterodimer with MDM2 and thereby enhance the MDM2 E3 ligase activity (Karni-Schmidt et al. 2016; Leslie and Zhang 2016). The MDM2–MDMX heterodimeric complex forms both through interactions between their respective RING domains (Leslie and Zhang 2016) and through short sequences at their respective C termini (Poyurovsky et al. 2007; Uldrijan et al. 2007). Additionally, through their respective abilities to bind to a region within the p53 transactivation domain, both MDM2 and MDMX can repress the ability of p53 to function as a transcription factor. The p53 protein itself activates the transcription of the MDM2 gene, thereby creating a negative feedback loop. The MDM2–MDMX heterodimer maintains low levels of p53 protein in unstressed cells, thereby reducing the amount of p53 available to alter the transcription of its targets (including MDM2) (Alarcon-Vargas and Ronai 2002). There are numerous hypotheses as to how MDM2 and MDMX regulate p53; the growing consensus is that they are necessary and nonredundant (Marine and Jochemsen 2004) for the regulation of p53. Mouse models (Wade et al. 2013) have shown that the two proteins have functions that can be both independent of, and dependent on, each other, based on context (Bielskienė et al VSports app下载. 2015; Karni-Schmidt et al. 2016).
MDM2 and MDMX also possess functions and interactions that are independent of p53. MDM2 in particular has been shown to have pro-oncogenic activities beyond regulating p53 (Bohlman and Manfredi 2014), and MDM2 can bind to a myriad of proteins, either altering their activity or causing their proteasome-mediated degradation (Fåhraeus and Olivares-Illana 2014) V体育官网. MDM2 has also been shown to regulate gene expression (Biderman et al. 2012; Jain and Barton 2016), heterochromatin rearrangement (Mungamuri et al. 2016; Wienken et al. 2017), DNA repair, and replication (Melo and Eischen 2012; Eischen 2017). Furthermore, this protein has roles in cellular metabolism (Riscal et al. 2016), EMT and metastasis (Wang et al. 2009; Chen et al. 2013; Lu et al. 2016; Chen et al. 2017a), cell survival and growth (Feeley et al. 2017), as well as mitochondrial dynamics (Arena et al. 2018). Although less studied, MDMX has also been shown to regulate cellular processes independently of p53. MDMX can enhance tumor progression in some cases by promoting cancer cell growth and genomic instability (Kadakia et al. 2002; Carrillo et al. 2015; Xiong et al. 2017). Supporting the idea that MDM2 and MDMX have p53-independent functions are data from human patients’ tumors that do not harbor wild-type p53, but that do have amplified levels of MDM2 or MDMX or both. It is also noteworthy that both MDM2 and MDMX can be activated by oncogenic stressors and mitogenic signals apart from p53 (Karni-Schmidt et al. 2016).
Intriguingly, in some settings MDMX may have tumor-suppressive roles in limiting cell transformation (Matijasevic et al. 2008) and tumor progression (Mancini et al. 2017), as well as promoting genome stability (Matijasevic et al. 2016). MDM2 has also been found to have tumor suppressor functions in addition to its oncogenic function (Manfredi 2010). For example, MDM2 has been reported to have growth-inhibitory domains (Deb 2002, 2003), which seem to be particularly manifested when MDM2 is ectopically expressed (Deb 2003; Ganguli and Wasylyk 2003).
Here, we report that MDM2 and MDMX promote ferroptosis in a manner that is independent of p53. These findings not only reveal two new key regulators of ferroptosis, they also provide a mechanism by which these proteins might exert tumor suppressor functions.
Results
p53 is not required for suppression of ferroptosis caused by inhibition of MDM2 (VSports最新版本)
We tested a number of human cancer cell lines for their sensitivity to erastin, a class I FIN that inhibits system Xc− activity. Consistent with prior reports, we found marked differences in viability of several cancer cell lines in response to this compound over a 24-h treatment period (Fig. 1A). For example, HCT116 colon cancer cells and H1299 lung cancer cells were virtually unaffected by erastin after 24 h of treatment. In contrast, both HT-1080 fibrosarcoma cells and SK-Hep1 liver cancer-derived cells were sensitive to erastin. Cell death in these erastin-sensitive cell lines was validated to be via ferroptosis, as it was reversed by ferrostatin-1 (fer-1), which inhibits lipid peroxidation and ferroptosis (Supplemental Fig. S1A; Dixon et al. 2012). When the time frame of the experiment was extended to 48 h, some cell death in HCT116 and H1299 cells was observed, which was reversed by fer-1 (Supplemental Fig. S1B), indicating that their death, while moderate and with a delayed onset, was via ferroptosis. Since there was not much difference in the response of HCT116 cells that harbor wild-type p53 and p53-null H1299 cells, we concluded that there was no obligate requirement for p53 to be present in its wild-type form for these cancer cells to undergo ferroptosis.
Figure 1.
p53 is not required for suppression of ferroptosis caused by inhibition of MDM2. (A) Varying responses of HT-1080, SK-Hep1, HCT116, and H1299 cells to erastin treatment. (B,C) Dose response of wild-type and p53 KO cells to erastin in HT-1080-derived cells (B) and SK-Hep1-derived cells (C). (D,E) Inhibition of erastin-induced HT-1080 cell death by Fer-1 (D) and DFO (E). (F–K) Effect of MDM2 antagonists (nutlin and MEL23) on the dose response of two different cell lines to erastin treatment. (F) Cartoon of known structure and mechanism of action of nutlin (Vassilev et al. 2004). (G,H) HT-1080-derived cells (G) and SK-Hep1-derived cells (H) treated with the combination of nutlin and erastin. (I) Cartoon of known structure and mechanism of action of MEL23 (Herman et al. 2011). (J,K) HT-1080-derived cells (J) and SK-Hep1-derived cells (K) treated with the combination of MEL23 and erastin. (L,M) Suppression of ferroptosis by the knockdown of MDM2 in two HT-1080-derived p53 KO clones. The top panel shows the viability of cells treated with a lethal dose of IKE when transfected with either the siRNA against Luciferase or one of two different siRNAs against MDM2. The bottom panel shows the corresponding decrease in the protein levels of MDM2 upon RNA interference against Luciferase (L) or MDM2 (1 and 2). The transfection was done using 15 nM of siRNA and the cells were treated with IKE 24 h after transfection. Cells in A–K were treated with drugs for 24 h. Cells in L and M were treated with drugs for 18 h. The data in A–E, G,H, J, and K represent the mean ± SE for two out of four independent experiments. The viability data in L and M represent the mean ± SE for four independent experiments. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to the DMSO control.
To determine the extent of ferroptosis as a function of p53 status in erastin-sensitive cancer cell lines, multiple clones of p53 knockout (KO) HT-1080 and SK-Hep1 cells were generated using CRISPR/Cas9 technology (Supplemental Fig. S1C). These cell lines were tested for their respective responses to a range of erastin concentrations (Fig. 1B,C). Consistent with previous reports (Murphy 2016), p53 KO derivatives were more resistant to erastin compared with their wild-type counterparts. Nevertheless, the HT-1080 and SK-Hep1 p53 KO clones were still more sensitive to erastin than were H1299 or HCT116 cells, and their death was reversed by fer-1, as well as by deferoxamine (DFO), which is an iron chelator that prevents ferroptosis (Fig. 1D,E; Supplemental Fig. S1D,E,H–K; Dixon et al. 2012). Neither fer-1 (Supplemental Fig. S1F,L) nor DFO (Supplemental Fig. S1G,M) had any effect on the responses of parental (WT) or p53 KO clones of HT-1080 and SK-Hep1 cells that were treated with staurosporine (STS), which elicits an apoptotic response (Belmokhtar et al. 2001). Thus, while in these cell lines p53 moderately sensitizes to ferroptosis, its presence is not required for ferroptosis.
Next, we evaluated the effects of two small molecule antagonists of MDM2: nutlin, which binds to the N-terminal region of MDM2 and blocks the primary site of the MDM2–p53 interaction (Fig. 1F; Vassilev et al. 2004), and MEL23 (MDM2 E3 ligase inhibitor 23), which blocks the E3 ligase activity of the MDM2–MDMX complex (Fig. 1I; Herman et al. 2011). Experiments with these compounds were complemented by the use of small interfering RNAs directed against MDM2 (see Fig. 1L,M; Supplemental Fig. S2K–S).
Unexpectedly, in the parental HT-1080 cells, nutlin treatment modestly reduced cell death by erastin (Fig. 1G), while MEL23 suppressed cell death induced by erastin even more effectively (Fig. 1J). This suggested that MDM2 might have a p53-independent function in facilitating erastin-induced ferroptosis. Indeed, in p53 KO cells, nutlin had little effect on cell death (Fig. 1G; Supplemental Fig. S2A,B), while, as was seen in the parental (WT) counterparts, MEL23 substantially decreased the extent of ferroptosis (Fig. 1J; Supplemental Fig. S2C,D). Similar results with MEL23 and nutlin were obtained in parental SK-Hep1 cells and SK-Hep1-derived p53 KO clones (Fig. 1H,K; Supplemental Fig. S2F–I). Note that MEL23 did not prevent apoptosis induced by STS treatment in any of these cell lines (Supplemental Fig. S2E,J), indicating that the effect of MEL23 does not extend to apoptosis.
The results with MEL23 suggested that MDM2 might promote erastin-mediated cell death with or without p53. Since MEL23 is an inhibitor of the MDM2–MDMX heterocomplex's E3 ligase activity, this indicated that MDM2 may have E3 ligase targets other than p53 that can promote ferroptosis. As nutlin functions to block the p53–MDM2 interaction, it would not necessarily be capable of inhibiting p53-independent functions of MDM2, perhaps explaining its inability to suppress erastin-induced cell death in p53 KO cells (insight into the suppression of ferroptosis by nutlin in parental cells expressing wild-type p53 is provided below).
We found that RNA interference against MDM2 in p53 KO HT-1080 cells also prevented ferroptosis. In these experiments, we induced ferroptosis with IKE, a more potent inhibitor of system Xc− (Fig. 1L,M). We obtained similar results by depleting MDM2 using RNAi in SK-Hep1-derived cells (Supplemental Fig. S2K,M,N,P,Q,S) As another control, ablation of MDM2 did not alter cell death upon treatment with STS (Supplemental Fig. S2L,O,R). In summary, these results demonstrate that MDM2 depletion or inhibition can selectively block ferroptosis, but not apoptosis, even in cancer cells lacking p53.
MDMX promotes sensitivity to ferroptosis independently of p53, alone and in complex with MDM2
We previously reported that MEL23 blocks the E3 ligase activity of the MDM2–MDMX heterodimer, preferentially over that of the MDM2 homodimer (Herman et al. 2011). Since MEL23 reduced sensitivity of cells to ferroptosis, we reasoned that lowering levels of the MDMX protein would reduce MDM2–MDMX complex formation and thereby might similarly suppress ferroptosis. We sought to deplete MDMX protein using the MDMX inhibitor NCS207895 (Berkson et al. 2005; Wang and Yan 2011), which represses expression from the MDMX promoter (Fig. 2A), and by using MDMX RNA interference, both of which result in lower MDMX protein levels. Indeed, treatment of cells with MEL23 or NCS207895, but not nutlin, prevented ferroptosis in both HT-1080 parental (Fig. 2B) and HT-1080-derived p53 KO (Fig. 2C; Supplemental Fig. S3A) cell lines. Similar results were obtained in SK-HEP1 parental (Supplemental Fig. S3B) and SK-HEP1-derived p53 KO cells (Supplemental Fig. S3C,D).
Figure 2.
MDMX promotes sensitivity to ferroptosis independently of p53. (A) Cartoon of the known structure and mechanism of action of the MDMX inhibitor (Berkson et al. 2005; Wang and Yan 2011). (B,C) Effect of MDM2 and MDMX antagonists (MEL23, nutlin, and MDMX inhibitor) on the lethality of erastin in HT-1080-derived cells. This effect was analyzed using microscopic images taken at 10× magnification (top panel) and cell viability (bottom left panel). (Bottom right panel) The corresponding MDM2/X protein levels were also measured. (D,E) Suppression of ferroptosis by the knockdown of MDMX in HT-1080-derived cells. The left panel shows the viability of cells treated with a lethal dose of IKE when transfected with either the scrambled siRNA negative control or one of two different siRNAs against MDMX. The right panel shows the corresponding decrease in the protein levels of MDMX upon RNA interference against the control (N) or MDMX (1 and 2). The transfection was done using 15 nM of siRNA and the cells were treated with IKE 24 h after transfection. Cells in B and C were treated with drugs for 16 h. Cells in D and E were treated with drugs for 18 h. The viability data in B and C represent the mean ± SE for two independent experiments. The viability data in D and E represent the mean ± SE for three independent experiments. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to the DMSO control.
Confirmation that MDMX is required for a ferroptotic response was obtained using experiments showing that either parental HT-1080/SK-Hep1 cells or their p53 KO derivatives underwent significantly less ferroptosis when depleted of MDMX by siRNAs (Fig. 2D,E; Supplemental Fig. S4A–D). Notably, the suppressive effects of MEL23 or siMDMX are not transient and do persist to a certain degree for at least 48 h after treatment (Supplemental Fig. S4E–G). Thus, MDMX is required for erastin-induced ferroptosis in these cell lines, and this function of MDMX does not require p53.
These data raised the question as to why nutlin treatment reduced ferroptosis in cells with wild-type p53 (Fig. 1; Supplemental Fig. S2). Consistent with previous reports (Patton et al. 2006; Wade et al. 2006; Xia et al. 2008), the levels of MDMX were markedly reduced upon nutlin treatment in the parental HT-1080 cells (Fig. 2B). In contrast, nutlin treatment did not reduce levels of MDMX in HT-1080 p53 KO cells (Fig. 2C; Supplemental Fig. S3A). Similarly, in parental SK-Hep1 cells (Supplemental Fig. S3B), nutlin treatment resulted in reduced MDMX protein abundance, which was not observed in p53 KO SK-Hep1 cells (Supplemental Fig. S3C,D). In contrast, and as expected, MEL23 led to accumulation of MDMX, as reported previously (Herman et al. 2011), by preventing its degradation by the MDM2 E3 ligase in both parental and p53 KO cells. Therefore, we hypothesize that the ability of nutlin to partially rescue viability in cells harboring wild-type p53 (but not KO cells that do not express p53) was due to its effect on the levels of MDMX protein. This finding, which suggests why nutlin was able to reduce ferroptosis in the presence of wild-type p53, provides further evidence that MDMX drives ferroptosis sensitivity.
The ability of MEL23 to suppress ferroptosis, in conjunction with the need for both MDM2 and MDMX to promote ferroptosis, led us to test whether the complex formed between MDM2 and MDMX mediates the ferroptosis sensitivity of cells, as opposed to the possibility that MDM2 and MDMX have independent functions. Towards this end, we ectopically expressed either wild-type MDMX or the MDMX variants MDMX (C463A) and MDMX (ΔC5), which lack the ability to bind to MDM2 (Supplemental Fig. S4H; Uldrijan et al. 2007; Huang et al. 2011). Overexpressed wild-type MDMX markedly increased cell death induced by erastin (Fig. 3) which could be suppressed by fer-1 (Supplemental Fig. S4I,J) in both HT-1080 and SK-Hep1 cells. This is consistent with the idea that MDMX is itself a target of the MDM2–MDMX complex and hence a limiting factor in the activity of the complex.
Figure 3.
Wild-type MDMX is more effective at sensitizing cells to ferroptosis than MDM2 binding-deficient mutants of MDMX. (A–D) Effect of overexpression of MDMX variants (WT and C463A) on the lethality of IKE in p53 KO clones derived from two different cells. The transfection was done using 300 µg of plasmid of each variant in HT-1080-derived cells (A) and SK-Hep1-derived cells (B). The transfection was done using a dose-curve (0–300 µg) of each variant in HT-1080-derived cells (C) and SK-Hep1-derived cells (D). The right panels of A and B and the bottom panels of C and D show the corresponding expression levels of the MDMX variants. The cells were treated for 18 h with IKE (5 µM) 24 h after transfection. The viability data in A–D represent the mean ± SE for three independent experiments. The viability after drug treatment has been measured using ATP-based CellTiter-Glo reagent and is normalized to the viability of the DMSO control under each transfection condition, respectively.
Overexpression of either mutant of MDMX produced significantly less ferroptosis, such that while both MDMX mutant proteins did cause an increase in cell death due to erastin, neither was as effective as wild-type MDMX despite being expressed at similar or even greater levels than wild-type MDMX (Fig. 3; Supplemental Fig. S4I,J). Attempts to overexpress MDM2 and perform comparable experiments were not technically successful.
These results indicate that the MDM2–MDMX complex plays a role in facilitating ferroptosis, but also suggest that MDMX may have additional functions beyond being a part of the MDM2–MDMX heterodimer. Together with the finding that MEL23 is known to inhibit the MDM2 E3 ligase mainly when in complex with MDMX, we infer a primary role for the MDM2–MDMX complex in promoting ferroptosis.
The MDM2–MDMX complex can promote ferroptosis in patient-derived glioblastoma models and rat brain slices (VSports app下载)
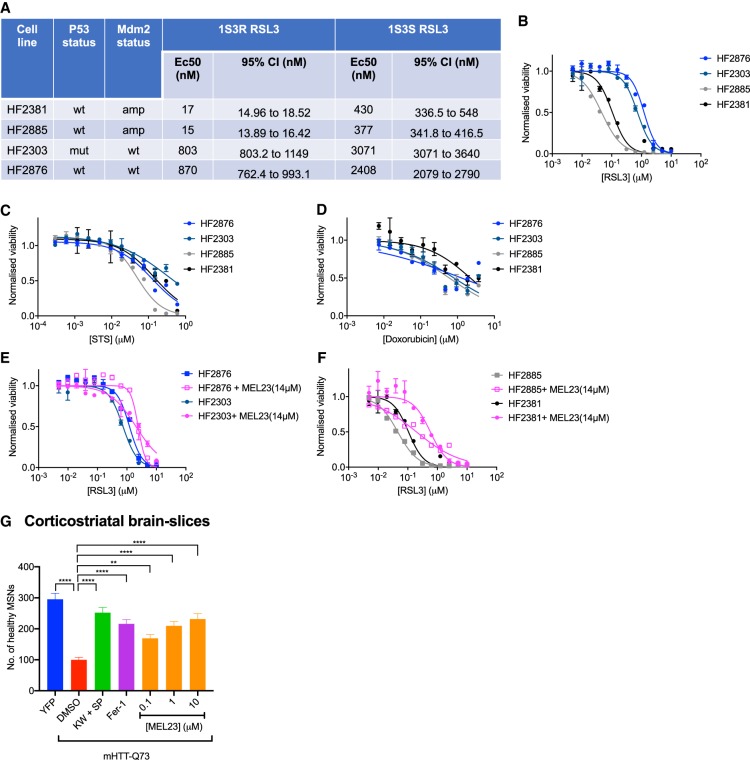
To examine whether MDM2 and MDMX can mediate ferroptosis in other contexts, we expanded our study to include patient-derived glioblastoma (GBM) cellular models, as well as an organotypic brain slice culture model of Huntington's disease. It was reported that patient glioblastoma models vary in their sensitivities to the ferroptosis inducer RSL3 (Quartararo et al. 2015). We compared the sensitivity of two GBM patient-derived models with high expression of MDM2 (Ye et al. 2020), but with wild-type TP53 versus two patient-derived cell models with normal expression level of MDM2 and wild-type or mutated TP53 (Fig. 4A). We found that these two sets of GBM models (high vs. normal expression of MDM2) had different responses to RSL3 (Fig. 4B); specifically, the models with high MDM2 expression had ∼50 times greater sensitivity to RSL3-induced ferroptosis. This trend in sensitivities of these models was specific to ferroptosis, as their response to apoptosis inducers, staurosporine, and doxorubicin, was similar (Fig. 4C,D). Notably, we were able to prevent cell death in both sets of GBM models with the coaddition of MEL23 (Fig. 4E,F). The models with high expression of MDM2 exhibited a stronger inhibition of RSL3-induced death upon cotreatment with MEL23, such that upon MEL23 treatment their response to RSL3 became similar to that of the wild-type MDM2-expressing models. These results suggest that MDM2, and likely the MDM2–MDMX complex, are able to drive the sensitivity of glioblastoma cells to ferroptosis.
Figure 4.
The MDM2–X complex can promote ferroptosis in patient-derived glioblastoma models and rat brain slices. (A–F) The MDM2–X complex mediates the sensitivity of four different patient-derived glioblastoma models to RSL3. (A) RSL3's EC50, MDM2, and p53 status for the glioblastoma models. Dose response of the glioblastoma models to RSL3 (B), staurosporine (STS) (C), and doxorubicin (D). MEL23 is able to inhibit the sensitivity of MDM2 wild-type glioblastoma models (E) and MDM2-amplified glioblastoma models (F) to RSL3 treatment. (G) MEL23 is able to suppress the mutant Huntingtin (mHTT) protein-induced neurodegeneration of rat striatal medium spiny neurons in a brain slice model of Huntington's disease. Rat corticostriatal brain slice explants were cotransfected with the first exon of the mHTT (Q73), and YFP transfection was used as a control. Brain slices were treated with either DMSO, a positive control mixture of 50 µM KW-6002 and 30 µM of SP600125, Fer-1 as a second positive control, or a three-point concentration response curve for MEL23. Cells in A–F were treated with drugs for 24 h. The data in A–F represent the mean ± SE for two out of four independent experiments. The data in G represent the mean ± SE from one of two representative experiments, with at least 24 samples assessed per condition. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to the DMSO control.
Next, we examined the role of the MDM2–MDMX complex in the medium spiny neurons (MSNs) of the striatum of rats using a postnatal brain slice model of Huntington's disease (Reinhart et al. 2011). These brain explants have been previously shown to lose a large portion of healthy MSNs when the first exon of mutant HTT (mHTT-Q73) is cotransfected with yellow fluorescent protein (YFP), as compared with the number of MSNs with YFP transfection alone (Reinhart et al. 2011; Kaplan et al. 2015). Since fer-1 can suppress ferroptotic death in this model (Skouta et al. 2014), we used it as a positive control to rescue the death of these MSNs with mutant HTT. We observed that MEL23 was also able to prevent mutant-HTT-induced death of these MSNs, even at low concentrations (Fig. 4G). This suggests that MEL23 is an inhibitor of ferroptosis in rat neurons, and that MDM2 and MDMX might mediate ferroptosis in the context of neurodegeneration.
Promotion of ferroptosis by MDM2 and MDMX is not dependent on the cell cycle
MDM2 was shown to ubiquitinate the p21 protein, thereby stimulating its degradation, both independently and in complex with MDMX (Warfel and El-Deiry 2013). Furthermore, p21 can have an inhibitory effect on ferroptosis in some contexts (Tarangelo et al. 2018). Therefore, we used p21 knockout (KO) clones that were derived from both HT-1080 and SK-Hep1 cells using the CRISPR/Cas9 system (Supplemental Fig. S5A) to show that the absence of p21 did not affect the degree of suppression of ferroptosis by MEL23 (Supplemental Fig. S5B,C).
Even in the absence of p53, MDM2 has been reported to promote cell growth and survival in some settings (Feeley et al. 2017), and these effects need not be dependent on p21. In line with this, we were unable to create stable MDM2 or MDMX knockout cells with CRISPR/Cas9 using either HT-1080 or SK-Hep1 p53 KO cells. This supports the notion that MDM2 and MDMX are necessary for cell survival in these cancer cell lines, even when lacking p53.
MEL23 and MDMX inhibitor were able to prevent ferroptotic cell death, even after 7–8 h of treatment (Supplemental Fig. S5D, left panel). At this time point, we did not find significant changes in the cell cycle that were common to the two MDM2–MDMX inhibiting compounds, which had similar effects on ferroptosis (Supplemental Fig. S5D, right panel). Taken together, these results indicate that the impact of blocking MDM2–MDMX complex activity on ferroptosis is independent of p21 and is not merely a by-product of cell cycle changes.
Inhibition of MDM2 and MDMX protects cells from ferroptosis caused by all classes of FINs
To gain insight into the role(s) of MDM2 and MDMX in promoting ferroptotic cell death, we aimed to identify which point in this process was affected by MDM2 and MDMX inhibition. This was accomplished by testing the impact of both MEL23 and the MDMX inhibitor on the response to other classes of FINs that induce ferroptosis through different mechanisms (Fig. 5A). Importantly, both compounds were able to suppress ferroptosis induced by all of the FINs, with the degree of rescue being the highest with class I FINs. Notably, these effects also did not require p53, although RSL3 was better suppressed in the presence of p53 (Fig. 5B; Supplemental Fig. S6A–C). Thus, MDM2 and MDMX were not limited to promoting ferroptosis induced by system Xc− inhibition; their roles extend to ferroptosis induced by all known mechanisms. We surmised that MDM2 and MDMX facilitate the lethal phospholipid peroxidation that is commonly affected by all four classes of FINs. Consistent with this conclusion, we confirmed that depletion of GSH, a characteristic of class I FINs, was not prevented by either of the MDM2 or MDMX inhibitors (Fig. 5C; Supplemental Fig. S6D). On the other hand, lipid peroxidation, which is a hallmark of all classes of FINs, was blocked by these same inhibitors (Fig. 5D; Supplemental Fig. S6E). This supports the likelihood that MDM2 and MDMX facilitate lipid peroxidation, a common requirement for all FINs.
Figure 5.
MDM2 and MDMX control a central checkpoint of ferroptosis. (A) Schematic depicting the previously established mechanisms of various FINs (Stockwell et al. 2017). (B) Inhibitory effect of MEL23 on the response of HT-1080-derived p53 KO cells to various classes of FINs: RSL3, FIN56, and FINO2. (C) Assessment of reduced glutathione levels in HT-1080-derived p53 KO cells treated with erastin with or without MEL23 or MDMX inhibitor. (D) Lipid ROS levels in HT-1080-derived p53 KO cells were measured by flow cytometry using C-11 Bodipy. The cells were treated with IKE in combination with either ferrostatin-1 or MDM2/X antagonists. The left panel represents the histogram showing the green fluorescence and the right panel represents the quantification of the normalized fluorescence. (E) The levels of labile (ferrous) iron in HT-1080-derived cells were measured using the FIP-1 probe. The cells were treated with erastin, either alone or in combination with DFO or MEL23. Cells in B were treated with drugs for 24 h. Cells in C–E were treated with erastin/IKE (10 µM) for 6.5 h. The data in B represent the mean ± SE for two out of four independent experiments. The data in C represent the mean ± SE for three biological replicates. The data in the right panel of D represent the mean ± SE for three independent experiments. The data in E is from one representative experiment out of three independent experiments. The data in E show the mean ± SE obtained from analyzing at least 100 different cells (from a minimum of five different fields) for each treatment condition. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to the DMSO control.
We examined the mechanism by which MDM2 and MDMX promote ferroptosis. We first focused on HT-1080 cells and their p53 KO derivatives, and used class I FINs to define the mechanism, as they showed the best suppression with MDM2–MDMX antagonists. All classes of FINs are dependent on the presence of labile iron, as iron chelators are able to block ferroptosis (Stockwell et al. 2017). Using FIP1, a cellular FRET-based iron probe, Aron et al. (2016) reported that erastin treatment causes an increase in the cellular levels of labile iron, which is brought back to basal levels by cotreatment with the iron chelator DFO. On the other hand, fer-1 can block ferroptosis without changing labile iron levels (compared with erastin treatment alone), demonstrating that reduction in iron levels is not the only means by which ferroptosis can be blocked (Aron et al. 2016). When FIP1 was used to test the impact of MDM2 and MDMX inhibition on iron levels, we found that, unlike DFO, neither MEL23 nor the MDMX inhibitor was able to lower the levels of labile iron in cells (Fig. 5E; Supplemental Fig. S6F). This led us to conclude that, in order to promote ferroptosis, MDM2 and MDMX do not need to alter iron levels in cells, and that these compounds do not act like iron chelators.
V体育安卓版 - MDM2 and MDMX prevent antioxidant responses during ferroptosis
MDM2 and MDMX alter the lipid metabolism of cells undergoing ferroptosis
Having ruled out the above mechanisms of suppressing ferroptosis, we performed untargeted lipidomic profiling to evaluate whether inhibition of MDM2 and MDMX changed the profile of lipids present in cells to disfavor lipid peroxidation. Cotreatment of cells with erastin and either MDM2 or MDMX inhibitors did indeed alter the lipid profile of cells. We observed two main trends in the lipid profile.
The first trend showed that these inhibitors caused an up-regulation of the abundance of many lipid species, including phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), and free fatty acids (FAs) (Fig. 6A; Supplemental Fig. S7A). This accumulation of lipids could indicate impaired catabolism, such as through β-oxidation. When there is incomplete β-oxidation, intermediate chain acyl carnitines (C5–C14) accumulate, while long chain acyl carnitines (C16–C22) are unaffected (Afshinnia et al. 2018), which we did not observe (Supplemental Fig. S7B,C). In addition to C5 and C16 carnitines, others such as C2 carnitines (derived from acetyl CoA obtained by metabolic breakdown products) and C4 carnitines (derived from both amino acid and lipid catabolism) (Koves et al. 2008) were increased in abundance upon MDM2–X inhibition. Additionally, when we blocked β-oxidation using etomoxir, which did indeed increase ferroptotic sensitivity as previously reported (Miess et al. 2018), there was no effect on the ability of MDM2/X antagonists to suppress ferroptosis (Supplemental Fig. S7D). Therefore, we concluded that altered β-oxidation is not the sole effect of MDM2 and MDMX inhibition. Instead, we assume that blocking these proteins led both to increased lipid anabolism and impaired lipid catabolism, and that the altered levels of acyl carnitines are simply markers of this global regulation of lipid metabolism.
Figure 6.
MDM2 and MDMX regulate lipid metabolism to favor ferroptosis. (A,B) Signal intensities of lipids (A) and triacyl glycerides (TAGs) (B) in HT-1080-derived p53 KO cells were measured by LC-MS and analyzed as significantly altered by ANOVA. The cells were treated with erastin in combination with MDM2 and MDMX antagonists. (C–F) The cell death due to erastin was assessed when PPARα activity was modulated using an agonist, pirinixic acid, or an antagonist, GW6471, in two different cell lines. Pirinixic acid blocks the lethal dose of erastin in HT-1080-derived p53 KO cells (C) and SK-Hep1-derived p53 KO cells (D). GW6471 enhances the dose response to erastin in HT-1080-derived p53 KO cells (E) and SK-Hep1-derived p53 KO cells (F). Cells in A and B were treated with erastin (10 µM) for 6.5 h. Cells in C and D were treated with erastin and pirinixic acid for 16 h. Cells in E and F were treated with erastin for 24 h and with GW6471 for 48 h. The data in A and B represent the mean ± SE for three biological replicates. The data in C and D represent the mean ± SE for three or two of three independent experiments, respectively. The data in E and F represent the mean ± SE for two out of four independent experiments. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to the DMSO control.
In the lipid profile analysis, we observed a second trend pertaining to triacylglycerides. In fact, both triacylglycerides and diacylglycerides were up-regulated upon erastin treatment, but were brought back to normal by the MDM2 and MDMX antagonists (Fig. 6B; Supplemental Fig. S7E). Blocking diacylglycerol acyltransferase (DGAT) to lower the triacyl glyceride levels directly was not able to block ferroptosis (Supplemental Fig. S7F,G). Therefore, the altered levels of triacylglycerides can also be viewed as a casualty of the altered lipid metabolism due to MDM2–X inhibition.
We hypothesized that cells treated with MDM2 and MDMX antagonists may up-regulate lipid metabolism pathways to combat oxidative stress. Among the up-regulated lipids upon MDM2–X inhibition were monounsaturated lipids (Fig. 6B), which were reported to be able to counteract ferroptosis (Magtanong et al. 2019). In a second line of evidence that is presented below, we also found the ability of MDM2–X to regulate another key component of antioxidant defense of cells, CoQ10.Taken together, these lipidomic data led us to consider the possibility that one or more master regulators of lipids might be under the control of MDM2 and MDMX.
PPARα activity plays a key role in facilitating the abilities of MDM2 and MDMX to promote ferroptosis
In order to identify the candidate master regulator, we first examined whether ferroptosis altered the localization of MDM2/X. We observed no consistent localization changes upon ferroptosis induction with erastin or suppression using DFO, MEL23, and MDMX inhibitor (Supplemental Fig. S8A). As an alternative, we examined previously known targets of MDM2 that have a potential link to ferroptosis. However, the knockdown of known targets of MDM2 with potential links to lipid metabolism (DHFR and FANCD2) did not prevent the activity of MDM2/X antagonists in ferroptosis (Supplemental Fig. S8B,C).
We then evaluated the role of PPARα, a transcription factor whose activity has been previously reported to be regulated by MDM2 under some conditions (Gopinathan et al. 2009). In fact, the PPAR family of transcription factors are known to be involved in large-scale rewiring of lipid homeostasis, particularly in response to stress (Gervois et al. 2000; van Raalte et al. 2004; Kersten 2008). PPARα is also reported to regulate carnitine palmitoyl transferases to promote the formation of acyl carnitines (Song et al. 2010; Chen et al. 2017b) and in fact, pharmacological fibrates are frequently used as triglyceride-lowering drugs that function by enhancing PPARα activity (Auwerx et al. 1996; van Raalte et al. 2004; Kersten 2008). Since we saw decreased abundance of triacylglycerides and increased abundance of acylcarnitines, in addition to large-scale changes to the lipid profile of cells treated with the MDM2 and MDMX antagonists, we hypothesized that this effect of MDM2 and MDMX on ferroptosis might be mediated by PPARα. To test this, we evaluated whether altering PPARα activity altered the sensitivity of cells to ferroptosis. To this end, we used both a PPARα-specific agonist (pirinixic acid) and a PPARα antagonist (GW6471). We confirmed the effectiveness of these compounds by testing their respective effects on select transcriptional targets of PPARα (Supplemental Fig. S9A,B). In the p53 KO derivatives of both HT-1080 and SK-Hep1 cells, increased PPARα activity suppressed ferroptosis (Fig. 6C,D), while decreased PPARα activity increased their sensitivity to ferroptosis (Fig. 6E,F). Akin to the effect of the PPARα agonist, the ectopic overexpression of PPARα also suppressed ferroptotic death (Supplemental Fig. S8D,E). On the other hand, PPARγ antagonist (GW9662) was able to decrease the ferroptotic sensitivity (Supplemental Fig. S9C,D) in line with reports indicating that the α and γ isoforms of PPAR have opposing cellular functions (Kersten 2008). We were unable to eliminate the PPARα protein using RNA interference or CRISPR technology.
Nevertheless, we tested whether MEL23 and MDMX inhibition were able to rescue ferroptosis when PPARα activity was reduced. Strikingly, in the absence of PPARα activity, neither MDM2–MDMX antagonist was effective in suppressing ferroptosis. As a control, the ability of fer-1 to inhibit ferroptosis was unaffected by the lack of PPARα activity (Fig. 7A–C; Supplemental Fig. S9E–G). We verified that the effects seen were not due to increased toxicity of compounds, such that the combination of either MEL23 or the MDMX inhibitor with the PPARα antagonist did not significantly decrease viability (Supplemental Fig. S9H,I), nor did cotreatment with the antagonist result in any changes in the ability of the antagonists to block or ablate MDM2 and MDMX (Fig. 7B,C; Supplemental Fig. S9E,G). Importantly, both MEL23 and the MDMX inhibitor themselves acted as agonists of PPARα activity, as seen by the up-regulation of known PPARα transcriptional targets (Fig. 7D; Supplemental Fig. S10A). This corroborates the published report that MDM2 can regulate PPARα activity (Gopinathan et al. 2009), and also suggests that the MDM2–MDMX complex is able to regulate this activity.
Figure 7.
PPARα activity plays a key role in facilitating the abilities of MDM2 and MDMX to dampen the antioxidant responses of cells and promote ferroptosis. (A,B) Effect of PPARα antagonist GW6471 on the ability of MEL23, MDMX inhibitor, and Fer-1 to block ferroptosis in SK-HEP1-derived p53 KO cells. (B, left panel) The relative cell death due to erastin when in combination with these inhibitors and GW6471. (B, right panel) The effect of MEL23 and MDMX inhibitor on the protein levels of MDM2 and MDMX is unaffected by cotreatment with GW6471. (A) Visualization of the aforementioned changes in cell viability at 10× magnification. (C) PPARα antagonist GW6471 suppresses the ability of siMDMX and siMDM2 to block ferroptosis in SK-HEP1-derived p53 KO cells. Two different siRNAs (#1 and #2) were used against each protein and compared against a scrambled siRNA negative control (siCtrl). The right panel shows the corresponding decrease in the protein levels of MDM2 and MDMX upon RNA interference. The transfection was done using 20 nM of siRNA in media with GW4671 and the cells were treated with drugs 24 h after transfection. (D) MEL23 and MDMX inhibitor up-regulate the mRNA levels of some known PPARα downstream targets in SK-HEP1-derived p53 KO cells. (E) Immunoprecipitation using PPARα antibody in SK-HEP1-derived p53 KO cells shows binding to MDM2 and MDMX even under treatment with MDM2–X antagonists. (F) Ratio of reduced to oxidized CoQ10 levels in HT-1080-derived p53 KO cells treated with erastin in combination with MDM2 and MDMX antagonists. (G) PPARα activity modulates the levels and extent of increase of FSP1 protein in cells treated with the MDM2/X antagonists in HT-1080 p53 KO cells. (H) Summary schematic depicting the hypothesized role of MDM2 and MDMX in ferroptosis. Cells in A–C, E, and G were treated with erastin/MEL23/MDMX inhibitor for 16 h and also with GW6471 for 40 h, as shown in A–C and G. Cells in D and F were treated with erastin (10 µM) for 6.5 h. The data in B and C represent the mean ± SE for three or two of three independent experiments, respectively. The data in D and F represent the mean ± SE for three biological replicates. The viability data have been measured using ATP-based CellTiter-Glo reagent and have been normalized to either the DMSO control or the GW6471 control, respectively, in B, and to each transfection's respective control that is not treated with erastin in C.
The exact mode of regulation of PPARα activity by MDM2–X is nonetheless unclear. These MDM2/X antagonists did not significantly or consistently alter the protein levels of PPARα (Supplemental Fig. S10B). Since PPARα is a transcription factor, nuclear exclusion could also alter its activity; we did not detect any change in localization of PPARα upon MDM2–X inhibition (Supplemental Fig. S10D), even in the context of ferroptosis (Supplemental Fig. S10C). This suggests that MDM2 and MDMX do not need to control the protein stability or nuclear localization of PPARα in order to regulate its transcriptional activity. On the other hand, both MDM2 and MDMX were detected when we immunoprecipitated PPARα, and this binding was not abrogated by the MDM2–X antagonists (Fig. 7E; Supplemental Fig. S10E,F). These results suggest that the MDM2–X complex may posttranslationally modify PPARα and alter its activity without inducing its degradation or nuclear exclusion.
MDM2/X alter the antioxidant responses of cells through PPARα activity
We evaluated whether heightened PPARα activity under MDM2–X antagonistic conditions could enhance other key defenses of cells. We measured the levels of CoQ10, a lipophilic antioxidant. In addition to its well-characterized role in the mitochondrial electron transport chain, CoQ10 can also suppress lipid peroxidation in mitochondrial and nonmitochondrial membranes (Shimada et al. 2016). Treatment with MDM2–X antagonists elicited a markedly increased ratio of reduced to oxidized CoQ10 during ferroptosis (Fig. 7F; Supplemental Fig. S10G). In fact, these inhibitors not only suppressed the reduction in the ratio resulting from erastin treatment, but enhanced it further beyond the levels of the vehicle control due to increased levels of reduced CoQ10 (Supplemental Fig. S10G, right panel). This finding is in line with our hypothesis of large-scale rewiring of lipid metabolism caused by MDM2 and MDMX to favor pro-oxidant cellular pathways.
Furthermore, it was reported recently that FSP1/AIFM2 can block ferroptosis independent of GPX4 by increasing the amount of reduced CoQ10 (Bersuker et al. 2019; Doll et al. 2019). We found that MDM2/X inhibition causes an increase in FSP1 levels, which likely in turn regenerates reduced CoQ10 to block ferroptosis. We also observed that in order to achieve the complete induction of FSP1 by MDM2–X inhibition, the full PPARα activity is required in either cell line (Fig. 7G; Supplemental Fig. S10H ).
These results thus indicate that lowered PPARα activity is a key conduit for MDM2/X to suppress the antioxidant defenses of cells and thereby promote ferroptosis. We provide a model in Fig. 7H depicting our proposed scheme of the roles of MDM2 and MDMX in preventing the normal ability of cancer cells to resist the lipid peroxidation that drives ferroptosis.
Discussion
Results obtained with small molecule inhibitors of MDM2 and MDMX, siRNA-mediated ablation of these proteins, and ectopically expressed MDMX variants, have revealed that MDM2 and MDMX can regulate ferroptosis in a p53-independent manner. Furthermore, these data show that MDM2 and MDMX control ferroptosis sensitivity through their ability to regulate lipid homeostasis. Blocking the MDM2–MDMX complex causes an accumulation of monounsaturated lipids and reduced CoQ10, as well as changes in acyl carnitines and tri/diacylglycerols. This indicates that inhibition of MDM2–MDMX complex activity leads to a coordinated rewiring of lipid metabolism to suppress ferroptosis. Of note, oleaginous microorganisms accumulate lipids in response to ROS (Shi et al. 2017), suggesting a similar stress response upon inhibition of MDMX and MDM2. The increase in reduced CoQ10 due to altered regulation of FSP1 has been shown to be a strong defense against lipid peroxidation (Bersuker et al. 2019; Doll et al. 2019). It is unlikely, however, that the increase in CoQ10 levels is the only means by which MDM2 and MDMX suppress ferroptosis, as MDM2 and MDMX inhibition also suppresses death induced by the class III FIN, FIN56, which can inhibit mevalonate-derived CoQ10 production (Shimada et al. 2016). It is therefore probable that additional defenses against ferroptosis are up-regulated in the absence of MDM2 and MDMX activity. Taken together, we assume that MDM2 and MDMX normally function to coordinate lipid metabolism, perhaps through one or more master regulators, in a manner that favors ferroptosis.
One such regulator is PPARα, the activity of which is modulated by the MDM2–MDMX antagonists and, in turn, this activity could mediate the role of MDM2 and MDMX in ferroptosis. Altered PPARα activity by MDM2 and MDMX disables the antioxidant defenses of cells, as well as effects other changes to the lipidome that together favor ferroptosis. The PPAR family of receptors, which are master regulators of cellular metabolism, particularly that of lipids, have been implicated in several diseases (Tyagi et al. 2011). PPARα activity in particular has been targeted to control hyperlipidemia (Auwerx et al. 1996), but of direct relevance here, it has also been implicated in controlling oxidative homeostasis of cells with implications in inflammation (van Raalte et al. 2004; Tyagi et al. 2011). It was reported that MDM2 can regulate the transcriptional activity of PPARα, (Gopinathan et al. 2009) and we found that, in these cell lines, both MDM2 and MDMX behave like PPARα antagonists. Since these results indicate that MDM2 and MDMX do not alter the PPARα protein levels, their inhibitory effect on PPARα is not due to its degradation. However, the results with MEL23, which is an inhibitor of the E3 ligase activity of the MDM2–MDMX complex, suggests either the possibility that blocking ubiquitination of PPARα (but not its degradation) affects its activity, or that degradation of a regulator of PPARα leads to its altered activity. Given that MDM2 and MDMX associate with PPARα, it is more likely that its activity is kept at bay through ubiquitination by the MDM2–X complex. Future studies will hopefully illuminate better how MDM2 and MDMX control PPARα in cells undergoing ferroptosis.
There are numerous other proteins that can potentially be modulated to render cells susceptible to ferroptosis (Yang and Stockwell 2016), either through PPARα or independently. For example, FSP1 protein levels were still induced, albeit to a lesser degree, even when PPARα activity was inhibited. It is possible that the MDM2–X complex can also directly control the stability of FSP1, along with regulating its transcription through PPARα or that there are other target proteins involved in this process. E3 ligase targets of MDM2, other than p53, have been discovered (Riley and Lozano 2012), but there are no confirmed targets of the MDM2–MDMX heterocomplex other than p53 and p21. Since we found that the role of the MDM2–MDMX complex in ferroptosis is independent of p53 and p21, other targets (as yet unknown) can likely mediate these effects on lipid metabolism. Our experiments indicated that some other known targets of MDM2 with ties to lipid metabolism (DHFR and FANCD2) are not involved. Identifying more targets of the MDM2–MDMX complex that can modulate the cellular lipid profile changes poses a substantial challenge for the future.
Our findings with MDMX overexpression suggest that MDMX may have additional MDM2-independent role(s) in promoting ferroptosis. It is possible that MDMX can coordinate additional changes in lipid metabolism, possibly through different master regulators. This is in agreement with a recent report showing that MDMX might control obesity in mice by preventing lipid accumulation (Kon et al. 2018).
Identifying a role for MDM2 and MDMX in ferroptosis may provide insight into therapeutic applications of ferroptosis inducers and inhibitors. The data presented here suggest that cancers with high levels of the MDM2–MDMX heterocomplex (and its E3 ligase activity) might be suitable targets for therapy involving ferroptosis inducers. Our results showing that the MDM2–MDMX complex can mediate the ferroptosis sensitivity of patient-derived glioblastoma cell lines further corroborates this hypothesis. There have been extensive efforts to develop nutlin-like compounds to treat cancers with wild-type p53 (Warner et al. 2012; Atatreh et al. 2018); one possible approach is to pair such compounds with other chemotherapeutics to achieve a more profound cancer control (Deben et al. 2015; Zanjirband et al. 2016). Given the discovery of p53-independent roles of MDM2 and MDMX, cancers with amplifications in MDM2 and MDMX (even in the absence of wild-type p53) (Wade et al. 2013; Pishas et al. 2015) might benefit from therapies that act as antagonists of MDM2 and MDMX. However, such therapies would likely not benefit from being combined with FINs or chemotherapeutics that induce ferroptosis, such as sorafenib (Lachaier et al. 2014) and sulfasalazine (Stockwell et al. 2017). With regard to such combinations, our data indicate that combining MDM2–MDMX antagonists with ferroptosis inducers would in fact be counterproductive. On the other hand, cancer therapies that activate p53 and cause unintended organ damage might be doing so through the induction MDM2/X mediated ferroptosis in those organs. For example, some studies suggest that the cardiotoxicity associated with doxorubicin treatment for breast cancer can be counteracted with CoQ10 supplementation (Conklin 2005). Such therapies might benefit from combinations with ferroptosis inhibitors to allow for higher doses of the therapy to be safely administered.
The results reported here could also aid in designing therapies for neurodegenerative disorders, ischemia, and organ damage, which have been shown to involve ferroptosis (Stockwell et al. 2017): MDM2–MDMX inhibitors might be used as inhibitors of degeneration in these diseases, as has been suggested for Fer-1 (Yang and Stockwell 2016). In support of this concept, we found that MEL23 has a robust neuroprotective effect in the brain slice model of Huntington's disease. MDM2 has also been shown to regulate kidney function, predominantly through its effects on p53, but also via p53-independent roles in modulating postischemic kidney injury (Mulay et al. 2012). Since there is mounting evidence that ferroptosis mediates kidney injury (Martin-Sanchez et al. 2017) and failure (Friedmann Angeli et al. 2014; Müller et al. 2017), our data provide additional support for the notion that MDM2 (and MDMX) antagonists might have potential in treating kidney dysfunction.
While most of this work was done in the absence of p53, we did find that the suppression of MDM2 and MDMX is effective in reducing the degree of ferroptosis in cells with functional p53. Especially in the context of the class II FIN RSL3, there seems to be a further desensitization to ferroptosis by inactivation of the MDM2–MDMX complex beyond the effect of loss of p53. We therefore suggest that MDM2 and MDMX may also be able to mediate the role of p53 in ferroptosis perhaps in different ways that are specific to particular classes of FINs. This mediatory role of the MDMs needs to be further evaluated to better understand the entirety of their role in ferroptosis.
Finally, MDM2 and MDMX may have context-dependent tumor suppressor functions in addition to their well described oncogenic functions mediated by inhibition of p53 (Manfredi 2010). Our results suggest a mechanism for this observation; namely, that MDM2 and MDMX can act as tumor suppressors by rewiring lipid metabolism in a manner that facilitates ferroptosis. Given that mounting evidence suggests that ferroptosis is a tumor-suppressive mechanism (Jiang et al. 2015; Jennis et al. 2016; Murphy 2016; Ou et al. 2016; Wang et al. 2016; Zhang et al. 2019), our data provide a new hypothesis for the field to explore.
"VSports" Materials and methods
Chemicals
The commercially available compounds used were erastin (Selleckchem S7242), nutlin-3a (Sigma-Aldrich 444152), MDMX inhibitor NSC207895 (Calbiochem 444158), staurosporine (Selleckchem S1421), deferoxamine (Calbiochem 252750), MEL23 (InterBioscreen) (Herman et al. 2011), PPARα agonist (Pirinixic acid; Selleckchem S8029), PPARα antagonist (GW6471; Santa Cruz Biotechnology CAS 436159-64-7), PPARγ antagonist (GW9662; Santa Cruz Biotechnology CAS 22978-25-2), and DGAT1 inhibitor (Cayman Chemicals A-922500).
The following compounds were synthesized: IKE as in Larraufie et al. (2015), fer-1 and RSL3 as in Dixon et al. (2012), and FIN56 as in Shimada et al. (2016).
All compounds were dissolved in DMSO (Sigma-Aldrich D8418). The fixed concentrations of compounds used was as follows (unless otherwise mentioned): 10 µM nutlin, 14 µM MEL23, 20 µM ferrostatin-1 (Fer-1), 90 µM deferoxamine (DFO), and 5 µM MDMX inhibitor.
"V体育安卓版" Cells
HCT116, H1299, and SK-Hep1 were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (Gemini Bioproducts 900-108). HT-1080 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (Gemini Bioproducts 900-108) and 1% nonessential amino acids (Sigma-Aldrich M7145). All parental cell lines were obtained from ATCC. The HT-1080 and SK-Hep1 parental cells were wild type for p53 and p21 and were edited using CRISPR technique (described below) to obtain p53 KO and p21 KO clones.
Cell viability assay (V体育2025版)
For dose response curves, 1800 cells were plated 36 µL per well of a 384-well plate on day 1. Compounds were dissolved in DMSO and a 12-point, twofold dilution series was prepared. The compounds were then diluted 1:33 in media and 4 µL was added to each well of the plates on day 2. After 24–48 h of treatment (depending on the cell line), the viability of cells was measured using 1:1 dilution of the CellTiter-Glo luminescent reagent (Promega G7573) with media, which was read on a Victor 5 plate reader after 10 min of shaking at room temperature. The intensity of luminescence was normalized to that of DMSO control. Some of the compounds tested were added at a constant concentration as specified to each dilution of the lethal compounds and corresponding amounts of DMSO was added to the control FIN treatment alone.
For siRNA experiments in SK-Hep1-derived cells, 1400 cells were plated for SK-Hep1 wild-type cells, while 1000 cells were plated for the SK-Hep1-derived p53 KO cells (clone 2 and clone 4). The rest of the viability assay procedure remained the same.
For viability assays performed in six-well plates, cells were harvested using trypsin (0.5 mL per well) and the medium was saved from each well. The trypsinized cells were resuspended with the collected medium and two to three aliquots (0.10 mL each) sampling different regions of this suspension were taken into 96-well plates to serve as technical replicates for the measurement. CellTiter-Glo luminescent viability assay was used to measure the viability of these aliquots. The rest of the culture was used to extract protein to be analyzed using Western blots.
Lipid profiling
Lipid profiling was performed as follows: For coenzyme Q10 detection and characterization using LC-MS, mass spectrometry-based targeted lipid profiling was done as described previously in Kraft et al. (2020). For lipid profile analysis used for estimation of lipids and acyl carnitines (University of Colorado Denver), mass spectrometry-based untargeted lipid profiling was done as described previously in Reisz et al. (2019). For lipid profile analysis used for estimation of triacyl and diacyl glycerols, mass spectrometry-based untargeted lipid profiling was done as described previously in Zhang et al. (2019).
Huntington brain slice assay (VSports在线直播)
Huntington brain slice assay was performed as described previously in Kaplan et al. (2015).
Statistical analysis
Prism (version 8, GraphPad) was used to make all the graphs in the paper and for performing all the statistical analysis shown. The GraphPad style (0.1234 [ns], <0.0332 [*], <0.0021 [**], and <0.0002 [***]) was used to represent the P-values. The P-values were calculated by ANOVA and appropriate multiple testing correction was done where required.
Standard procedures were used for genome editing, immunoblot, immunoprecipitation, quantitative reverse transcription PCR, transfection, flow cytometry, and measurement of reduced glutathione, labile iron, and lipid ROS. Please see the Supplemental Material for details.
Competing interest statement
C.P. is a member of the Scientific Advisory Board of Aileron Therapeutics. B.R.S. is a consultant to and has equity in Inzen Therapeutics. B.R.S. also is an inventor on patents and patent applications related to ferroptosis.
Supplementary Material
V体育官网入口 - Acknowledgments
We are grateful to Ella Freulich for technical assistance; Joshua Choe, Chen Katz, and Vitalay Fomin for advice; and Yan Zhang, Michael Gaschler, and Rachid Skouta for synthesizing compounds (RSL3, IKE, fer-1, and FIN56) used in this study. We also thank Jiandong Chen for supplying the mAb against MDMX. This study was supported by CA87497 to B.R.S. and C.P., R35CA220526 to C.P., and R35CA209896 and R61NS109407 to B.R.S. This study also benefited from R01GM79465 to C.J.C.; A.T.A. thanks the National Science Foundation for a graduate fellowship and was partially supported by Chemical Biology Training Grant T32GM066698.
Author contributions: D.V. designed and conducted the majority of the experiments in the paper under the guidance of C.P. and B.R.S. D.V., C.P., and B.R.S. wrote and edited the manuscript. N.A.O. designed and conducted several experiments mainly relating to PPARα. F.Z. measured the abundance of coenzyme Q10 and other lipids. D.R.T. created the p53 KO cells and also attempted to create MDM2 and MDMX KO cells. M.E.S. designed and created the CRISPR/Cas9 viruses for the p21 KO cells, and independently validated the effects of MDM2 and MDMX inhibition on ferroptosis. D.E.D. and D.C.L. were responsible for the corticostriatal brain slice assay. E.S.K. was responsible for conducting experiments shown in Supplemental Figure S5B under D.V.’s guidance. A.T.A. and C.J.C. helped set up the iron probe experiments and also synthesized the probe. A.M.K. performed the cell cycle analysis. J.M.C. optimized and aided in performing the GSH estimation assays. S.-H.M. isolated the SK-Hep1-derived p21 KO cells. M.C. provided the FSP1 antibody. A.D.’s group measured the abundance of lipids and acyl carnitines. N.A.O., D.R.T., F.Z., M.E.S., and A.D. wrote the Materials and Methods subsections.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.334219.119.
References
- Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, Hawkins J, Bellovich K, Gipson D, Michailidis G, et al. 2018. Impaired β-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J Am Soc Nephrol 29: 295–306. 10.1681/ASN.2017030350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon-Vargas D, Ronai Z. 2002. p53-Mdm2—the affair that never ends. Carcinogenesis 23: 541–547. 10.1093/carcin/23.4.541 [V体育2025版 - DOI] [PubMed] [Google Scholar]
- Arena G, Cissé MY, Pyrdziak S, Chatre L, Riscal R, Fuentes M, Arnold JJ, Kastner M, Gayte L, Bertrand-Gaday C, et al. 2018. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol Cell 69: 594–609.e8. 10.1016/j.molcel.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AT, Loehr MO, Bogena J, Chang CJ. 2016. An endoperoxide reactivity-based FRET probe for ratiometric fluorescence imaging of labile iron pools in living cells. J Am Chem Soc 138: 14338–14346. 10.1021/jacs.6b08016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atatreh N, Ghattas MA, Bardaweel SK, Al Rawashdeh S, Al Sorkhy M. 2018. Identification of new inhibitors of Mdm2–p53 interaction via pharmacophore and structure-based virtual screening. Drug Des Devel Ther 12: 3741–3752. 10.2147/DDDT.S182444 [VSports注册入口 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Schoonjans K, Fruchart JC, Staels B. 1996. Regulation of triglyceride metabolism by PPARs: fibrates and thiazolidinediones have distinct effects. J Atheroscler Thromb 3: 81–89. 10.5551/jat1994.3.81 [DOI (V体育安卓版)] [PubMed] [Google Scholar]
- Belmokhtar CA, Hillion J, Ségal-Bendirdjian E. 2001. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 20: 3354–3362. 10.1038/sj.onc.1204436 [DOI] [PubMed] [Google Scholar]
- Berkson RG, Hollick JJ, Westwood NJ, Woods JA, Lane DP, Lain S. 2005. Pilot screening programme for small molecule activators of p53. Int J Cancer 115: 701–710. 10.1002/ijc.20968 [DOI] [PubMed] [Google Scholar]
- Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al. 2019. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575: 688–692. 10.1038/s41586-019-1705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biderman L, Manley JL, Prives C. 2012. Mdm2 and MdmX as regulators of gene expression. Genes Cancer 3: 264–273. 10.1177/1947601912455331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielskienė K, Bagdonienė L, Mozūraitienė J, Kazbarienė B, Janulionis E. 2015. E3 ubiquitin ligases as drug targets and prognostic biomarkers in melanoma. Medicina 51: 1–9. 10.1016/j.medici.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Bohlman S, Manfredi JJ. 2014. p53-independent effects of Mdm2. Subcell Biochem 85: 235–246. 10.1007/978-94-017-9211-0_13 [DOI (VSports app下载)] [PMC free article] [PubMed] [Google Scholar]
- Carrillo AM, Bouska A, Arrate MP, Eischen CM. 2015. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 34: 846–856. 10.1038/onc.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qiu J, Yang D, Lu J, Yan C, Zha X, Yin Y. 2013. MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PLoS One 8: e78794 10.1371/journal.pone.0078794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang D-D, Wu Y-P, Su D, Zhou T-Y, Gai R-H, Fu Y-Y, Zheng L, He Q-J, Zhu H, et al. 2017a. MDM2 promotes epithelial–mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer 117: 1192–1201. 10.1038/bjc.2017.265 [V体育2025版 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Huang Y, Zeng H, Hu B, Guan L, Zhang H, Yu A-M, Johnson CH, Gonzalez FJ, et al. 2017b. PPARα regulates tumor cell proliferation and senescence via a novel target gene carnitine palmitoyltransferase 1C. Carcinogenesis 38: 474–483. 10.1093/carcin/bgx023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin KA. 2005. Coenzyme Q10 for prevention of anthracycline-induced cardiotoxicity. Integr Cancer Ther 4: 110–130. 10.1177/1534735405276191 [V体育2025版 - DOI] [PubMed] [Google Scholar]
- Deb SP. 2002. Function and dysfunction of the human oncoprotein mdm2. Front. Biosci 7: d235–d243. 10.2741/A723 [DOI (V体育平台登录)] [PubMed] [Google Scholar]
- Deb SP. 2003. Cell cycle regulatory functions of the human oncoprotein MDM21 1 NIH (CA74172 and CA70712). Mol Cancer Res 1: 1009–1016. [PubMed] [Google Scholar]
- Deben C, Wouters A, de Beeck KO, van Den Bossche J, Jacobs J, Zwaenepoel K, Peeters M, Van Meerbeeck J, Lardon F, Rolfo C, et al. 2015. The MDM2-inhibitor Nutlin-3 synergizes with cisplatin to induce p53 dependent tumor cell apoptosis in non-small cell lung cancer. Oncotarget 6: 22666–22679. 10.18632/oncotarget.4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. 2012. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149: 1060–1072. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Grocin AG, da Silva TNX, Panzilius E, Scheel CH, et al. 2019. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575: 693–698. 10.1038/s41586-019-1707-0 [DOI] [PubMed] [Google Scholar]
- Eischen CM. 2017. Role of Mdm2 and Mdmx in DNA repair. J Mol Cell Biol 9: 69–73. 10.1093/jmcb/mjw052 ["V体育ios版" DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus R, Olivares-Illana V. 2014. MDM2's social network. Oncogene 33: 4365–4376. 10.1038/onc.2013.410 [DOI] [PubMed] [Google Scholar]
- Feeley KP, Adams CM, Mitra R, Eischen CM. 2017. Mdm2 is required for survival and growth of p53-deficient cancer cells. Cancer Res 77: 3823–3833. 10.1158/0008-5472.CAN-17-0809 ["VSports app下载" DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. 2014. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16: 1180–1191. 10.1038/ncb3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B. 2003. p53-independent functions of MDM2. Mol Cancer Res 1: 1027–1035. [VSports最新版本 - PubMed] [Google Scholar]
- Gervois P, Inés PT, Fruchart J-C, Staels B. 2000. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin Chem Lab Med 38: 3–11. 10.1515/CCLM.2000.002. [DOI (V体育官网入口)] [PubMed] [Google Scholar]
- Gopinathan L, Hannon DB, Peters JM, Vanden Heuvel JP. 2009. Regulation of peroxisome proliferator-activated receptor-α by MDM2. Toxicol Sci 108: 48–58. 10.1093/toxsci/kfn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman AG, Hayano M, Poyurovsky MV, Shimada K, Skouta R, Prives C, Stockwell BR. 2011. Discovery of Mdm2–MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov 1: 312–325. 10.1158/2159-8290.CD-11-0104 [VSports最新版本 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yan Z, Liao X, Li Y, Yang J, Wang Z-G, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. 2011. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci 108: 12001–12006. 10.1073/pnas.1102309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Barton MC. 2016. Outside the p53 RING: transcription regulation by chromatin-bound MDM2. Mol Cell 62: 805–807. 10.1016/j.molcel.2016.05.035 [VSports app下载 - DOI] [PubMed] [Google Scholar]
- Jennis M, Kung C-P, Basu S, Budina-Kolomets A, Leu JI-J, Khaku S, Scott JP, Cai KQ, Campbell MR, Porter DK, et al. 2016. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev 30: 918–930. 10.1101/gad.275891.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang S-J, Su T, Hibshoosh H, Baer R, Gu W. 2015. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520: 57–62. 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadakia M, Brown TL, McGorry MM, Berberich SJ. 2002. MdmX inhibits Smad transactivation. Oncogene 21: 8776–8785. 10.1038/sj.onc.1205993 [DOI] [PubMed] [Google Scholar]
- Kaplan A, Gaschler MM, Dunn DE, Colligan R, Brown LM, Palmer AG, Lo DC, Stockwell BR. 2015. Small molecule-induced oxidation of protein disulfide isomerase is neuroprotective. Proc Natl Acad Sci 112: E2245–E2252. 10.1073/pnas.1500439112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni-Schmidt O, Lokshin M, Prives C. 2016. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol 11: 617–644. 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. 2008. Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Res 2008: 132960 10.1155/2008/132960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Wang D, Tongyuan Li LJ, Qiang L, Gu W. 2018. Inhibition of Mdmx (Mdm4) in vivo induces anti-obesity effects. Oncotarget 9: 7282–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JRB, Newgard CB, et al. 2008. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 7: 45–56. 10.1016/j.cmet.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Kraft VAN, Bezjian CT, Pfeiffer S, Ringelstetter L, Müller C, Zandkarimi F, Merl-Pham J, Bao X, Anastasov N, Kössl J, et al. 2020. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent Sci 6: 41–53. doi:10.1021/acscentsci.9b01063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaier E, Louandre C, Godin C, Saidak Z, De Picardie U, Verne J, Cedex A. 2014. Sorafenib induces ferroptosis in human cancer cell lines originating from different solid tumors. Anticancer Res 34: 6417–6422. [PubMed] [Google Scholar]
- Larraufie M-H, Yang WS, Jiang E, Thomas AG, Slusher BS, Stockwell BR. 2015. Incorporation of metabolically stable ketones into a small molecule probe to increase potency and water solubility. Bioorg Med Chem Lett 25: 4787–4792. 10.1016/j.bmcl.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie PL, Zhang Y. 2016. MDM2 oligomers: antagonizers of the guardian of the genome. Oncogene 35: 6157–6165. 10.1038/onc.2016.88 [DOI (VSports最新版本)] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Yan C, Huang Y, Shi D, Fu Z, Qiu J, Yin Y. 2016. Mouse double minute 2 (MDM2) upregulates Snail expression and induces epithelial-to-mesenchymal transition in breast cancer cells in vitro and in vivo. Oncotarget 7: 37177–37191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ko P-J, To M, Cao JY, Forcina GC, Tarangelo A, Ward CC, Cho K, Patti GJ, Nomura DK, et al. 2019. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem Biol 26: 420–432.e9. 10.1016/j.chembiol.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Teveroni E, Di Conza G, Monteleone V, Arisi I, Pellegrino M, Buttarelli M, Pieroni L, D'Onofrio M, Urbani A, et al. 2017. MDM4 actively restrains cytoplasmic mTORC1 by sensing nutrient availability. Mol Cancer 16: 55 10.1186/s12943-017-0626-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi JJ. 2010. The Mdm2–p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24: 1580–1589. 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J-C, Jochemsen AG. 2004. Mdmx and Mdm2: brothers in arms? Cell Cycle 3: 898–902. 10.4161/cc.3.7.998 [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez D, Ruiz-Andres O, Poveda J, Carrasco S, Cannata-Ortiz P, Sanchez-Niño MD, Ruiz Ortega M, Egido J, Linkermann A, Ortiz A, et al. 2017. Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid-induced AKI. J Am Soc Nephrol 28: 218–229. 10.1681/ASN.2015121376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijasevic Z, Steinman HA, Hoover K, Jones SN. 2008. MdmX promotes bipolar mitosis to suppress transformation and tumorigenesis in p53-deficient cells and mice. Mol Cell Biol 28: 1265–1273. 10.1128/MCB.01108-07 ["VSports app下载" DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijasevic Z, Krzywicka-Racka A, Sluder G, Gallant J, Jones SN. 2016. The Zn-finger domain of MdmX suppresses cancer progression by promoting genome stability in p53-mutant cells. Oncogenesis 5: e262 10.1038/oncsis.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo AN, Eischen CM. 2012. Protecting the genome from mdm2 and mdmx. Genes Cancer 3: 283–290. 10.1177/1947601912454139 [VSports注册入口 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Miess H, Dankworth B, Gouw AM, Rosenfeldt M, Schmitz W, Jiang M, Saunders B, Howell M, Downward J, Felsher DW, et al. 2018. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 37: 5435–5450. 10.1038/s41388-018-0315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay SR, Thomasova D, Ryu M, Anders H-J. 2012. MDM2 (murine double minute-2) links inflammation and tubular cell healing during acute kidney injury in mice. Kidney Int 81: 1199–1211. 10.1038/ki.2011.482 [DOI] [PubMed] [Google Scholar]
- Müller T, Dewitz C, Schmitz J, Schröder AS, Bräsen JH, Stockwell BR, Murphy JM, Kunzendorf U, Krautwald S. 2017. Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell Mol Life Sci 74: 3631–3645. 10.1007/s00018-017-2547-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungamuri SK, Qiao RF, Yao S, Manfredi JJ, Gu W, Aaronson SA. 2016. USP7 enforces heterochromatinization of p53 target promoters by protecting SUV39H1 from MDM2-mediated degradation. Cell Rep 14: 2528–2537. 10.1016/j.celrep.2016.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ME. 2016. Ironing out how p53 regulates ferroptosis. Proc Natl Acad Sci 113: 12350–12352. 10.1073/pnas.1615159113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Wang S-J, Li D, Chu B, Gu W. 2016. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci 113: E6806–E6812. 10.1073/pnas.1607152113 ["VSports手机版" DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. 2006. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res 66: 3169–3176. 10.1158/0008-5472.CAN-05-3832 [DOI (VSports最新版本)] [PubMed] [Google Scholar]
- Pishas KI, Adwal A, Neuhaus SJ, Clayer MT, Farshid G, Staudacher AH, Callen DF. 2015. XI-006 induces potent p53-independent apoptosis in Ewing sarcoma. Sci Rep 5: 11465 10.1038/srep11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KLB, Pan Z-Q, Pavletich N, Prives C. 2007. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 26: 90–101. 10.1038/sj.emboj.7601465 ["V体育官网" DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartararo CE, Reznik E, deCarvalho AC, Mikkelsen T, Stockwell BR. 2015. High-throughput screening of patient-derived cultures reveals potential for precision medicine in glioblastoma. ACS Med Chem Lett 6: 948–952. 10.1021/acsmedchemlett.5b00128 [DOI (VSports)] [PMC free article] [PubMed] [Google Scholar]
- Reinhart PH, Kaltenbach LS, Essrich C, Dunn DE, Eudailey JA, DeMarco CT, Turmel GJ, Whaley JC, Wood A, Cho S, et al. 2011. Identification of anti-inflammatory targets for Huntington's disease using a brain slice-based screening assay. Neurobiol Dis 43: 248–256. 10.1016/j.nbd.2011.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz JA, Zheng C, D'Alessandro A, Nemkov T. 2019. Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics. Methods Mol Biol 1978: 121–135. 10.1007/978-1-4939-9236-2_8 [DOI] [PubMed] [Google Scholar]
- Riley MF, Lozano G. 2012. The many faces of MDM2 binding partners. Genes Cancer 3: 226–239. 10.1177/1947601912455322 [VSports - DOI] [PMC free article] [PubMed] [Google Scholar]
- Riscal R, Schrepfer E, Arena G, Cissé MY, Bellvert F, Heuillet M, Rambow F, Bonneil E, Sabourdy F, Vincent C, et al. 2016. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol Cell 62: 890–902. 10.1016/j.molcel.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Shi K, Gao Z, Shi T-Q, Song P, Ren L-J, Huang H, Ji X-J. 2017. Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: the state of the art and future perspectives. Front Microbiol 8: 793 10.3389/fmicb.2017.00793 [DOI (VSports在线直播)] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, Brown LM, Valenzuela CA, Wolpaw AJ, Stockwell BR. 2016. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol 12: 497–503. 10.1038/nchembio.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, Rosenberg PA, Lo DC, Weinberg JM, Linkermann A, et al. 2014. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc 136: 4551–4556. 10.1021/ja411006a ["VSports最新版本" DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Attia RR, Connaughton S, Niesen MI, Ness GC, Elam MB, Hori RT, Cook GA, Park EA. 2010. Peroxisome proliferator activated receptor α (PPARα) and PPARγ coactivator (PGC-1α) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol Cell Endocrinol 325: 54–63. 10.1016/j.mce.2010.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al. 2017. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171: 273–285. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarangelo A, Magtanong L, Bieging-Rolett KT, Li Y, Ye J, Attardi LD, Dixon SJ. 2018. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep 22: 569–575. 10.1016/j.celrep.2017.12.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S. 2011. The peroxisome proliferator-activated receptor: a family of nuclear receptors role in various diseases. J Adv Pharm Technol Res 2: 236–240. 10.4103/2231-4040.90879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek W-J, Vousden KH. 2007. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 26: 102–112. 10.1038/sj.emboj.7601469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raalte DH, Li M, Haydn Pritchard P, Wasan KM. 2004. Peroxisome proliferator-activated receptor (PPAR)-α: a pharmacological target with a promising future. Pharm Res 21: 1531–1538. 10.1023/B:PHAM.0000041444.06122.8d [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. 2006. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem 281: 33036–33044. 10.1074/jbc.M605405200 [DOI] [PubMed] [Google Scholar]
- Wade M, Li Y-C, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature Reviews Cancer 13: 83–96. 10.1038/nrc3430 [DOI (V体育安卓版)] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yan C. 2011. A small-molecule p53 activator induces apoptosis through inhibiting MDMX expression in breast cancer cells. Neoplasia 13: 611–619. 10.1593/neo.11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-P, Wang W-L, Chang Y-L, Wu C-T, Chao Y-C, Kao S-H, Yuan A, Lin C-W, Yang S-C, Chan W-K, et al. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11: 694–704. 10.1038/ncb1875 [DOI] [PubMed] [Google Scholar]
- Wang S-J, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, Gu W. 2016. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep 17: 366–373. 10.1016/j.celrep.2016.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfel Na, El-Deiry WS. 2013. p21WAF1 and tumourigenesis. Curr Opin Oncol 25: 52–58. 10.1097/CCO.0b013e32835b639e [DOI] [PubMed] [Google Scholar]
- Warner WA, Sanchez R, Dawoodian A, Li E, Momand J. 2012. Identification of FDA-approved drugs that computationally bind to MDM2. Chem Biol Drug Des 80: 631–637. 10.1111/j.1747-0285.2012.01428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken M, Moll UM, Dobbelstein M. 2017. Mdm2 as a chromatin modifier. J Mol Cell Biol 9: 74–80. 10.1093/jmcb/mjw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Knezevic D, Tovar C, Huang B, Heimbrook DC, Vassilev LT. 2008. Elevated MDM2 boosts the apoptotic activity of p53-MDM2 binding inhibitors by facilitating MDMX degradation. Cell Cycle 7: 1604–1612. 10.4161/cc.7.11.5929 [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, et al. 2017. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep 20: 1692–1704. 10.1016/j.celrep.2017.07.055 [DOI] [PubMed] [Google Scholar]
- Xiong S, Pant V, Zhang Y, Aryal NK, You MJ, Kusewitt D, Lozano G. 2017. The p53 inhibitor Mdm4 cooperates with multiple genetic lesions in tumourigenesis. J Pathol 241: 501–510. 10.1002/path.4854 [V体育安卓版 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. 2016. Ferroptosis: death by lipid peroxidation. Trends Cell Biol 26: 165–176. 10.1016/j.tcb.2015.10.014 [DOI (VSports)] [PMC free article] [PubMed] [Google Scholar]
- Ye LF, Reznik E, Korn JM, Lin F, Yang G, Malesky K, Gao H, Loo A, Pagliarini R, Mikkelsen T, et al. 2020. Patient-derived glioblastoma cultures as a tool for small-molecule drug discovery. Oncotarget 11: 443–451. 10.18632/oncotarget.27457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanjirband M, Edmondson RJ, Lunec J. 2016. Pre-clinical efficacy and synergistic potential of the MDM2–p53 antagonists, Nutlin-3 and RG7388, as single agents and in combined treatment with cisplatin in ovarian cancer. Oncotarget 7: 40115–40134. 10.18632/oncotarget.9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhuang L, Gan B. 2019. BAP1 suppresses tumor development by inducing ferroptosis upon SLC7A11 repression. Mol Cell Oncol 6: 1536845 10.1080/23723556.2018.1536845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.