VSports - MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling
- PMID: 32079652
- PMCID: PMC7111265
- DOI: 10.1101/gad.334219.119
MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling
Abstract
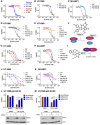
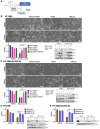
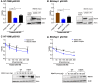
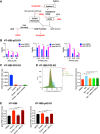
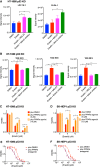
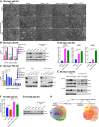
MDM2 and MDMX, negative regulators of the tumor suppressor p53, can work separately and as a heteromeric complex to restrain p53's functions. MDM2 also has pro-oncogenic roles in cells, tissues, and animals that are independent of p53. There is less information available about p53-independent roles of MDMX or the MDM2-MDMX complex. We found that MDM2 and MDMX facilitate ferroptosis in cells with or without p53 VSports手机版. Using small molecules, RNA interference reagents, and mutant forms of MDMX, we found that MDM2 and MDMX, likely working in part as a complex, normally facilitate ferroptotic death. We observed that MDM2 and MDMX alter the lipid profile of cells to favor ferroptosis. Inhibition of MDM2 or MDMX leads to increased levels of FSP1 protein and a consequent increase in the levels of coenzyme Q10, an endogenous lipophilic antioxidant. This suggests that MDM2 and MDMX normally prevent cells from mounting an adequate defense against lipid peroxidation and thereby promote ferroptosis. Moreover, we found that PPARα activity is essential for MDM2 and MDMX to promote ferroptosis, suggesting that the MDM2-MDMX complex regulates lipids through altering PPARα activity. These findings reveal the complexity of cellular responses to MDM2 and MDMX and suggest that MDM2-MDMX inhibition might be useful for preventing degenerative diseases involving ferroptosis. Furthermore, they suggest that MDM2/MDMX amplification may predict sensitivity of some cancers to ferroptosis inducers. .
Keywords: CoQ10; FSP1; MDM2; MDMX; PPARα; cancer; ferroptosis; lipid metabolism; p53-independent V体育安卓版. .
© 2020 Venkatesh et al V体育ios版. ; Published by Cold Spring Harbor Laboratory Press. .
Figures (VSports)

References
-
- Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, Hawkins J, Bellovich K, Gipson D, Michailidis G, et al. 2018. Impaired β-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J Am Soc Nephrol 29: 295–306. 10.1681/ASN.2017030350 - DOI (V体育ios版) - PMC - PubMed
-
- Alarcon-Vargas D, Ronai Z. 2002. p53-Mdm2—the affair that never ends. Carcinogenesis 23: 541–547. 10.1093/carcin/23.4.541 - DOI (VSports手机版) - PubMed
-
- Aron AT, Loehr MO, Bogena J, Chang CJ. 2016. An endoperoxide reactivity-based FRET probe for ratiometric fluorescence imaging of labile iron pools in living cells. J Am Chem Soc 138: 14338–14346. 10.1021/jacs.6b08016 - "V体育官网入口" DOI - PMC - PubMed
-
- Atatreh N, Ghattas MA, Bardaweel SK, Al Rawashdeh S, Al Sorkhy M. 2018. Identification of new inhibitors of Mdm2–p53 interaction via pharmacophore and structure-based virtual screening. Drug Des Devel Ther 12: 3741–3752. 10.2147/DDDT.S182444 - "VSports注册入口" DOI - PMC - PubMed
Publication types
MeSH terms
- "V体育2025版" Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- VSports在线直播 - Actions
- V体育2025版 - Actions
"V体育安卓版" Substances
- VSports app下载 - Actions
- VSports - Actions
- VSports在线直播 - Actions
- "VSports最新版本" Actions
- "VSports最新版本" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous (V体育平台登录)
