T Cell Recruitment to the Intestinal Stem Cell Compartment Drives Immune-Mediated Intestinal Damage after Allogeneic Transplantation
- PMID: 31278057
- PMCID: PMC7239328
- DOI: "VSports最新版本" 10.1016/j.immuni.2019.06.003
T Cell Recruitment to the Intestinal Stem Cell Compartment Drives Immune-Mediated Intestinal Damage after Allogeneic Transplantation
Abstract
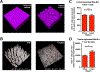
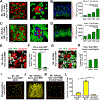
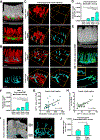
The key sites within the gastrointestinal (GI) tract where T cells mediate effector responses and the impact of these responses on intestinal stem cells (ISCs) remain unclear. Using experimental bone marrow transplantation to model immune-mediated GI damage and 3D imaging to analyze T cell localization, we found that the ISC compartment is the primary intestinal site targeted by T cells after transplantation. Recruitment to the crypt base region resulted in direct T cell engagement with the stem cell compartment and loss of crypt base columnar ISCs, which expressed both MHC classes I and II VSports手机版. Vasculature expressing the adhesion molecule MAdCAM-1 clustered near the crypt base, preferentially regulating crypt compartment invasion and ISC reduction without affecting T cell migration to villi. These findings indicate that allogeneic T cells rapidly access the stem cell niche after transplantation, and this targeted recruitment to the stem cell compartment results in ISC loss during immune-mediated GI damage. .
Keywords: BMT; GVHD; ISCs; LPAM; MAdCAM-1; Paneth cells; allogeneic bone marrow transplantation; beta7 integrin; graft versus host disease; imaging of immunity; intestinal stem cells; mucosal immunology; transplantation. V体育安卓版.
Copyright © 2019 Elsevier Inc V体育ios版. All rights reserved. .
Figures




References
-
- Adams DH, and Eksteen B (2006). Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat Rev Immunol 6, 244–251. - PubMed
-
- Agudo J, Park ES, Rose SA, Alibo E, Sweeney R, Dhainaut M, Kobayashi KS, Sachidanandam R, Baccarini A, Merad M, and Brown BD (2018). Quiescent Tissue Stem Cells Evade Immune Surveillance. Immunity 48, 271–285 e275. - "V体育2025版" PMC - PubMed
-
- Barker N (2014). Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15, 19–33. - PubMed
Publication types
- "VSports app下载" Actions
- Actions (V体育安卓版)
MeSH terms
- "VSports注册入口" Actions
- Actions (V体育安卓版)
- VSports在线直播 - Actions
- "VSports手机版" Actions
- Actions (V体育官网)
- "V体育2025版" Actions
- V体育ios版 - Actions
- "VSports注册入口" Actions
- "V体育2025版" Actions
Substances
Grants and funding
LinkOut - more resources (V体育官网入口)
Full Text Sources
V体育安卓版 - Medical
"VSports注册入口" Molecular Biology Databases
VSports在线直播 - Research Materials

