Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4+ memory T cells
- PMID: 31118448
- PMCID: PMC6531457
- DOI: 10.1038/s41467-019-10018-1
Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4+ memory T cells
Abstract (VSports手机版)
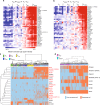
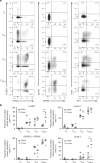
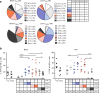
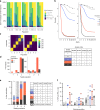
All memory T cells mount an accelerated response on antigen reencounter, but significant functional heterogeneity is present within the respective memory T-cell subsets as defined by CCR7 and CD45RA expression, thereby warranting further stratification. Here we show that several surface markers, including KLRB1, KLRG1, GPR56, and KLRF1, help define low, high, or exhausted cytokine producers within human peripheral and intrahepatic CD4+ memory T-cell populations VSports手机版. Highest simultaneous production of TNF and IFN-γ is observed in KLRB1+KLRG1+GPR56+ CD4 T cells. By contrast, KLRF1 expression is associated with T-cell exhaustion and reduced TNF/IFN-γ production. Lastly, TCRβ repertoire analysis and in vitro differentiation support a regulated, progressive expression for these markers during CD4+ memory T-cell differentiation. Our results thus help refine the classification of human memory T cells to provide insights on inflammatory disease progression and immunotherapy development. .
"V体育安卓版" Conflict of interest statement
The authors declare no competing interests.
VSports app下载 - Figures



References
Publication types
"V体育平台登录" MeSH terms
- Actions (V体育安卓版)
- V体育ios版 - Actions
- "V体育2025版" Actions
- "V体育平台登录" Actions
- Actions (V体育平台登录)
- "V体育2025版" Actions
- Actions (V体育平台登录)
Substances
- "V体育2025版" Actions
Grants and funding
- e:Kid/Bundesministerium für Bildung, Wissenschaft und Kultur (Federal Ministry of Education, Science and Culture)/International
- SFB650/Deutsche Forschungsgemeinschaft (German Research Foundation)/International
- e:kid/Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (Federal Ministry for Education, Science, Research and Technology)/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

