Comparison of Salmonella enterica Serovars Typhi and Typhimurium Reveals Typhoidal Serovar-Specific Responses to Bile
- PMID: 29229736
- PMCID: PMC5820949
- DOI: 10.1128/IAI.00490-17
Comparison of Salmonella enterica Serovars Typhi and Typhimurium Reveals Typhoidal Serovar-Specific Responses to Bile
Abstract
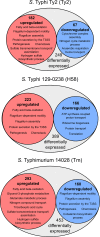
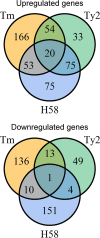
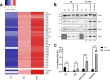
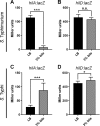
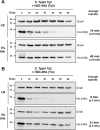
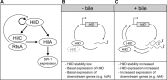
Salmonella enterica serovars Typhi and Typhimurium cause typhoid fever and gastroenteritis, respectively VSports手机版. A unique feature of typhoid infection is asymptomatic carriage within the gallbladder, which is linked with S Typhi transmission. Despite this, S Typhi responses to bile have been poorly studied. Transcriptome sequencing (RNA-Seq) of S Typhi Ty2 and a clinical S Typhi isolate belonging to the globally dominant H58 lineage (strain 129-0238), as well as S Typhimurium 14028, revealed that 249, 389, and 453 genes, respectively, were differentially expressed in the presence of 3% bile compared to control cultures lacking bile. fad genes, the actP-acs operon, and putative sialic acid uptake and metabolism genes (t1787 to t1790) were upregulated in all strains following bile exposure, which may represent adaptation to the small intestine environment. Genes within the Salmonella pathogenicity island 1 (SPI-1), those encoding a type IIII secretion system (T3SS), and motility genes were significantly upregulated in both S Typhi strains in bile but downregulated in S Typhimurium. Western blots of the SPI-1 proteins SipC, SipD, SopB, and SopE validated the gene expression data. Consistent with this, bile significantly increased S Typhi HeLa cell invasion, while S Typhimurium invasion was significantly repressed. Protein stability assays demonstrated that in S Typhi the half-life of HilD, the dominant regulator of SPI-1, is three times longer in the presence of bile; this increase in stability was independent of the acetyltransferase Pat. Overall, we found that S Typhi exhibits a specific response to bile, especially with regard to virulence gene expression, which could impact pathogenesis and transmission. .
Keywords: H58 clade; RNA-Seq; SPI-1 regulation; bile responses; cell invasion; typhoid fever. V体育安卓版.
Copyright © 2018 Johnson et al.
V体育平台登录 - Figures

References
-
- Dougan G, Baker S. 2014. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol 68:317–336. doi:10.1146/annurev-micro-091313-103739. - VSports app下载 - DOI - PubMed
-
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12:117–124. doi:10.1016/j.mib.2008.12.001. - "VSports最新版本" DOI - PMC - PubMed
-
- Altier C. 2005. Genetic and environmental control of Salmonella invasion. J Microbiol 43:85–92. - PubMed
Publication types
- V体育平台登录 - Actions
- Actions (VSports最新版本)
MeSH terms
- "VSports手机版" Actions
- VSports - Actions
- V体育安卓版 - Actions
- Actions (V体育官网)
- "VSports在线直播" Actions
- "V体育ios版" Actions
Substances (V体育官网)
Grants and funding
"VSports app下载" LinkOut - more resources
"VSports手机版" Full Text Sources
Other Literature Sources
Medical
Miscellaneous (VSports注册入口)

