Global intron retention mediated gene regulation during CD4+ T cell activation
- PMID: 27369383
- PMCID: PMC5001615
- DOI: 10.1093/nar/gkw591
Global intron retention mediated gene regulation during CD4+ T cell activation
Abstract
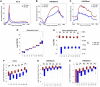
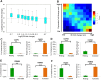
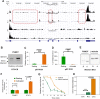
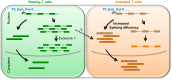
T cell activation is a well-established model for studying cellular responses to exogenous stimulation VSports手机版. Using strand-specific RNA-seq, we observed that intron retention is prevalent in polyadenylated transcripts in resting CD4(+) T cells and is significantly reduced upon T cell activation. Several lines of evidence suggest that intron-retained transcripts are less stable than fully spliced transcripts. Strikingly, the decrease in intron retention (IR) levels correlate with the increase in steady-state mRNA levels. Further, the majority of the genes upregulated in activated T cells are accompanied by a significant reduction in IR. Of these 1583 genes, 185 genes are predominantly regulated at the IR level, and highly enriched in the proteasome pathway, which is essential for proper T cell proliferation and cytokine release. These observations were corroborated in both human and mouse CD4(+) T cells. Our study revealed a novel post-transcriptional regulatory mechanism that may potentially contribute to coordinated and/or quick cellular responses to extracellular stimuli such as an acute infection. .
Published by Oxford University Press on behalf of Nucleic Acids Research 2016 V体育安卓版. This work is written by (a) US Government employee(s) and is in the public domain in the US. .
V体育2025版 - Figures



References (V体育官网)
-
- Chandok M.R., Farber D.L. Signaling control of memory T cell generation and function. Semin. Immunol. 2004;16:285–293. - PubMed
-
- Macian F., Lopez-Rodriguez C., Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. - VSports注册入口 - PubMed
-
- Bendfeldt H., Benary M., Scheel T., Steinbrink K., Radbruch A., Herzel H., Baumgrass R. IL-2 Expression in Activated Human Memory FOXP3(+) Cells Critically Depends on the Cellular Levels of FOXP3 as Well as of Four Transcription Factors of T Cell Activation. Front. Immunol. 2012;3:264. - PMC - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
VSports手机版 - MeSH terms
- V体育2025版 - Actions
- "VSports手机版" Actions
- "VSports注册入口" Actions
- "V体育2025版" Actions
- V体育安卓版 - Actions
- "V体育官网" Actions
- "V体育ios版" Actions
- VSports注册入口 - Actions
- VSports - Actions
- "V体育官网入口" Actions
VSports - Substances
- "V体育平台登录" Actions
- "VSports" Actions
Grants and funding
LinkOut - more resources (VSports手机版)
Full Text Sources
Other Literature Sources
Research Materials

