V体育官网 - Dual and Opposite Effects of hRAD51 Chemical Modulation on HIV-1 Integration
- PMID: 26051216
- PMCID: V体育安卓版 - PMC4889029
- DOI: 10.1016/j.chembiol.2015.04.020
Dual and Opposite Effects of hRAD51 Chemical Modulation on HIV-1 Integration (V体育平台登录)
Abstract
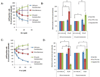
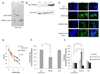
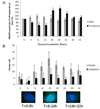
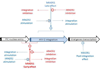
The cellular DNA repair hRAD51 protein has been shown to restrict HIV-1 integration both in vitro and in vivo. To investigate its regulatory functions, we performed a pharmacological analysis of the retroviral integration modulation by hRAD51 VSports手机版. We found that, in vitro, chemical activation of hRAD51 stimulates its integration inhibitory properties, whereas inhibition of hRAD51 decreases the integration restriction, indicating that the modulation of HIV-1 integration depends on the hRAD51 recombinase activity. Cellular analyses demonstrated that cells exhibiting high hRAD51 levels prior to de novo infection are more resistant to integration. On the other hand, when hRAD51 was activated during integration, cells were more permissive. Altogether, these data establish the functional link between hRAD51 activity and HIV-1 integration. Our results highlight the multiple and opposite effects of the recombinase during integration and provide new insights into the cellular regulation of HIV-1 replication. .
Copyright © 2015 Elsevier Ltd. All rights reserved V体育安卓版. .
Figures



V体育2025版 - References
-
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. - PubMed
Publication types
- V体育官网入口 - Actions
MeSH terms
- VSports在线直播 - Actions
- VSports - Actions
- Actions (VSports app下载)
- "VSports手机版" Actions
- "VSports在线直播" Actions
- V体育官网入口 - Actions
- "VSports注册入口" Actions
- "VSports注册入口" Actions
- VSports手机版 - Actions
- "VSports app下载" Actions
- VSports app下载 - Actions
- Actions (V体育安卓版)
- VSports最新版本 - Actions
Substances
- Actions (VSports最新版本)
- "VSports在线直播" Actions
- "VSports" Actions
- V体育安卓版 - Actions
- V体育安卓版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

