SUMMARY
The cellular DNA repair hRAD51 protein has been shown to restrict HIV-1 integration both in vitro and in vivo. To investigate its regulatory functions, we performed a pharmacological analysis of the retroviral integration modulation by hRAD51. We found that, in vitro, chemical activation of hRAD51 stimulates its integration inhibitory properties, whereas inhibition of hRAD51 decreases the integration restriction, indicating that the modulation of HIV-1 integration depends on the hRAD51 recombinase activity. Cellular analyses demonstrated that cells exhibiting high hRAD51 levels prior to de novo infection are more resistant to integration VSports最新版本. On the other hand, when hRAD51 was activated during integration, cells were more permissive. Altogether, these data establish the functional link between hRAD51 activity and HIV-1 integration. Our results highlight the multiple and opposite effects of the recombinase during integration and provide new insights into the cellular regulation of HIV-1 replication.
INTRODUCTION
Retroviral replication requires the integration of the viral cDNA into the host cell genome, a multistep process catalyzed by the intasome complex formed between the retroviral integrase (IN) and the viral DNA [1, 2]. After intasome binding to the host chromatin and insertion of the viral cDNA ends into the target DNA the post-integration repair (PIR) of the integration locus occurs V体育平台登录. This step, probably catalyzed by host factors, leads to the stable insertion of the viral genome, the provirus (for reviews, see [3–5]). We have previously shown that hRAD51, belonging to the homologous recombination DNA repair pathway (HR), could bind HIV-1 IN [6] and exert a negative effect on integration both in vitro and in vivo [7]. Indeed, the stimulation of hRAD51 activity by the RAD stimulatory compound 1 (RS-1 [8]), inhibits HIV-1 integration, leading to a significant decrease of the viral replication [7]. Moreover, hRAD51 has also been shown to activate the HIV-1 LTR dependent-transcription [9–11] and, thus, to stimulate viral genes expression. Taken together, these data indicate that hRAD51 plays different roles during the retroviral replication cycle that must be taken into account by clinical approaches. To better understand hRAD51 regulatory functions we performed a pharmacological analysis of the impact of hRAD51 chemical modulation on HIV-1 integration. Our results showed that hRAD51 stimulatory and inhibitory compounds induced multiple and opposite effects on this specific step of the retroviral replication cycle. These data indicate that efficient integration relies on equilibrium between pro- and anti-integration properties of hRAD51. This suggests that cellular pathways and/or treatments affecting this equilibrium could differently affect viral replication and reveals the complex regulatory functions of hRAD51 on integration.
RESULTS
Selection of hRAD51 chemical modulators affecting its HIV-1 integrase inhibition ability
hRAD51 inhibits HIV-1 integration in vitro (SI1) and enhancement of the hRAD51/DNA active nucleofilaments formation by the RAD51 stimulatory compound 1, RS-1, (chemical structure shown in figure 1A) improves the integration inhibition by the recombinase both in vitro and in vivo [7]. These data highlighted the role of the nucleocomplex in the inhibition process. To better characterize the mechanism of inhibition, we searched for compounds capable of affecting the hRAD51/DNA binding properties and, thus, the formation of the active nucleofilament. Stilbenes, like DIDS, having previously been shown to affect hRAD51 activity [12], we tested natural stilbenes derivatives recently identified in our laboratory [13] in a hRAD51/DNA interaction assay. As reported in SI2, most of the 22 molecules tested were found to inhibit the hRAD51/DNA association but only one, the E-pterostilbene (P-ter) (chemical structure shown in figure 1A), stimulated binding. Given that hRAD51 stimulatory compounds are attractive candidates as antiviral agents [7], we first focused our interest on the stimulatory molecule. To verify its possible stimulation effect on hRAD51 strand exchange activity, P-ter was assayed in an in vitro recombination activity. As shown in SI3A, P-ter stimulates the recombinase activity at least as efficiently as RS-1, (EC50 for P-ter = 19 VSports注册入口. 2 ± 2. 4 µM and EC50 for RS-1 = 25 ± 3 µM).
Figure 1. Effect of hRAD51 modulators on HIV-1 integration.
The chemical structure of the stimulatory compounds RS-1 and P-ter (A) and the inhibitory compounds RI-1 and DIDS, as well as the sequence of the aptamers (B) are indicated. Increasing concentrations of wt-hRAD51 were added in a standard concerted integration assay in the presence of 100 µM ATP (w/o molecule or aptamer), and with 7. 5 or 15 µM RS-1 (C), P-ter (D), RI-1 (E) or 0. 1, 0 V体育官网入口. 25 or 0. 5 µM of A30 (F). The data reported represent the mean values of at least three independent experiments ± standard deviation (error bars). The activity in the absence of compounds, corresponding to the total amount of donor DNA integrated into the acceptor plasmid, as detected on agarose gel electrophoresis and shown in SI1, was normalized to 100 %.
The hRAD51 stimulatory compounds were then tested on in vitro hRAD51 mediated inhibition of HIV-1 IN activity using the concerted integration assay described in SI1. As reported in SI4A and [13], P-ter was previously shown to inhibit HIV-1 IN activity with an IC50 = 47. 5 ± 2 VSports在线直播. 5 µM. Since P-ter showed hRAD51 stimulatory effect between 5–20 µM (see SI3) without significantly affecting HIV-1 IN activity, it was tested at concentrations up to 15 µM on hRAD51-mediated IN inhibition. As reported in figure 1C, a strong improvement in integration restriction was observed in the presence of P-ter, as observed in the case of RS-1 (figure 1D). This indicates that the stimulation of the hRAD51 activity not only by RS-1 but also by other stimulatory compounds as P-ter can enhance the HIV-1 IN inhibition properties of the recombinase. To determine whether the active hRAD51 nucleofilament could serve as a target for modulating hRAD51 mediated inhibition of HIV-1 IN, we next examined whether inhibition of the recombinase could also affect its integration restriction properties. Two hRAD51 chemical inhibitors: DIDS [12] and RI-1 [14, 15] and several DNA aptamers previously selected against hRAD51, [16] and figure 1B, were tested. These molecules were assayed on the in vitro hRAD51 recombination activity shown in SI3A. Both RI-1 and DIDS displayed an inhibitory activity on the strand exchange catalyzed by hRAD51 (IC50 determined for RI-1 and DIDS were respectively 50 ± 5 µM and 90 ± 2. 5 µM, SI3B). A47 and A30 aptamers [16] were also found to strongly inhibit hRAD51 under these conditions (IC50 for A47 = 75 ± 4 nM and IC50 for A30 = 30 ± 2 nM), while their shortened control versions A47c and A30c did not (SI3C). Among the hRAD51 inhibitory molecules assayed, DIDS showed a significant IN inhibitory effect (IC50 = 5 ± 2 µM, SI4B) and, thus, was excluded from further analyses. We next compared the effect of RS-1 and P-ter with that of RI-1 and A30, which showed the best inhibitory effect, on hRAD51-mediated integration restriction. As reported in figure 1E and F good correlation was observed between hRAD51 activity and IN inhibition. Indeed, all the hRAD51 inhibitors induced a significant decrease in the hRAD51-mediated integration inhibition, which is in sharp contrast to the potentiation observed with the hRAD51 stimulatory compounds RS-1 and P-ter. No effect of molecules on the IN/hRAD51 interaction was detected (SI5) confirming that the modulation of hRAD51-mediated inhibition by the drugs was mainly due to their effect on the active hRAD51 nucleofilaments. This suggested that modulators of the nucleocomplex could be used as tools to explore its biological regulatory function in infected cells.
Opposite effects of hRAD51 chemical modulation on HIV-1 integration step in 293T cells
RS-1 has previously been shown to stimulate hRAD51 activity both in vitro and in vivo [8] and to inhibit HIV-1 replication in single- and multiple-round infection assays performed in different cell types including primary PBMC resting cells [7]. To better characterize the mechanism of action of RS-1, especially at the integration step, we compared it to P-ter using a typical 293T single-round replication assay, in order to focus on the early steps of infection V体育2025版. Cytotoxicity measurement in cells treated with increasing concentrations of drug showed no significant effect on cells viability (CC50 > 250 µM, SI6A and B). We next tested the drugs for their effect on cellular hRAD51-mediated DNA repair activity using a typical cisplatin resistance assay in a non-toxic concentration range. As reported in SI7A and B, a 24-hour treatment of 293T cells with either RS-1 or P-ter induced an increase in the cisplatin resistance, as expected from the stimulation of the active hRAD51 nucleofilament. To determine the effect of the molecule on intracellular hRAD51 protein, we performed an immunolocalization analysis of the recombinase. As reported in SI7C, cells treated with either RS-1or P-ter showed an increased number of hRAD51 nuclear foci as compared to untreated cells. Quantification of the cytoplasmic and nuclear foci (SI7D) confirmed that treatment with the hRAD51 stimulatory compounds induced a nuclear relocalization of hRAD51, consistent with a stimulation of the formation of active nucleofilaments in the nuclear compartment. Since i) hRAD51 activity could be altered during cell cycle and ii) the effect of the compounds on the viral replication might depend on the alternation of cell cycle we analyzed the impact of drugs treatments on the cellular cycle. As reported in SI7E, propidium iodide labeling of the cells showed no significant change in the cell cycle alternation of the treated versus untreated cells. This allowed further analyzes of drugs effects on early steps of retroviral replication.
As shown in figure 2A a 24-hour treatment of 293T cells with RS-1 prior to transduction with pNL4. 3 based pRRLsin-PGK-eGFP-WPRE VSV-G pseudo-typed viruses induced an inhibition of transduction efficiency. Quantification of the different viral DNA populations indicated that this phenotype was due to an inhibition of integration, as shown by an increase of the amount of unintegrated two-LTR DNA circles, and a decrease of the integrated DNA form, while the total DNA amount remained unchanged (figure 2B). In contrast, treatments performed 5 hours after transduction induced an opposite phenotype, showing a stimulation of the viral replication correlated with an increased integration. To determine whether this dual effect was specific to the RS-1 molecule, we tested the newly selected P-ter. Assays performed on 293T cells transduced with the lentiviral vector produced results similar to those obtained with RS-1 (figure 2C and 2D) VSports.
Figure 2. Effect of RS-1 and P-ter treatment on early steps of HIV-1 replication and viral DNA populations.
Cells were treated for 24 hours prior to, concomitantly, or 5–16 hours post-transduction with either RS-1 (A and B) or P-ter (C and D). The eGFP fluorescence was measured 10 days post-transduction by flow cytometry. The percentage of eGFP-positive cells in the absence of compound was normalized to 100% (A and C). The amount of the total, integrated and 2-LTR circles viral DNA forms was quantified as described in Materials and Methods at a fixed 30 µM concentration (effective and non-cytotoxic) of RI-1 or p-ter, under two distinct treatment conditions (early and late). The amount of each viral DNA species produced in the absence of compound was normalized to 100 % (B and D). Results are the mean of three independent experiments. The p-values calculated using a Student’s t-test are indicated as *p<0.05, **p<0.005.
The RI-1 compound, exerting an opposite effect to RS-1 on hRAD51, was then tested. As reported in SI6C, no significant RI-1 toxicity was observed in a 1–50 µM range, though a slight decrease in cell viability could be observed at concentrations higher than 50 µM (CC50 = 200 ± 15 µM). A 24-hour pre-treatment of 293T cells with RI-1 was found to induce a decrease in both the cisplatin resistance (SI8A) and the nuclear foci formation (SI8B). These results confirmed that RI-1 could negatively modulate the hRAD51 DNA repair activity. The drug was next tested on early steps of retroviral replication. As reported in figure 3A, a 24-hour pre-treatment led to an increase of the percentage of eGFP-positive 293T cells transduced with the lentiviral vector (EC50 = 50 ± 12 µM). Quantification of the viral DNA species indicated that the phenotype was due to a stimulation of integration, as shown by the significant increase in integrated DNA forms and the decrease of two-LTR circles, the global viral DNA amount remaining unchanged (figure 3B). Strikingly, a 5-hour post-transduction treatment with RI-1 induced an opposite effect. Indeed, the viral replication was inhibited (EC50 = 50 ± 6 µM), with a decrease in the integrated DNA forms accompanied by slight increase of the two-LTR circles. In contrast, a 16-hour post-transduction treatment had no significant effect on replication.
Figure 3. Effect of hRAD51 inhibitory compounds RI-1 on early steps of HIV-1 replication.
Cells were treated for 24 hours prior to, concomitantly, or 5–16 hours post-transduction. eGFP fluorescence was measured 10 days following transduction by flow cytometry. The percentage of GFP-positive cells obtained in the absence of compound was normalized to 100 % (A). The effect of RI-1 on viral DNA production was measured by quantifying the total, integrated and 2-LTR circles viral DNA forms at a fixed 50 µM concentration (non-cytotoxic and effective) of RI-1, under two treatment conditions. The proportion of the different DNA species obtained in the absence of compound was normalized to 100 % (B). Results are represented as the mean values calculated from three independent experiments. The p-values are reported as *p<0.05, **p<0.005.
These results demonstrated that the hRAD51 activity modulation can have opposite effects on integration depending on the chronology of the treatment. Especially, stimulation of the recombinase prior to transduction rendered the cells resistant to integration, whereas stimulation of hRAD51 post-transduction had a positive effect on integration. Thus, depending on its catalytic status, hRAD51 can either positively or negatively influence the early steps of HIV-1 replication by acting at the integration step. In view of these data, one important conclusion is that cells with higher hRAD51 intracellular concentrations would be expected to be more resistant to HIV-1 infection. To verify this hypothesis, we studied the effect of the hRAD51 intracellular concentration on integration and viral replication.
Modulation of hRAD51 expression affects both cellular DNA repair activity and HIV-1 integration
In order to increase the hRAD51 intracellular content, a hRAD51-FLAG tagged protein was overexpressed in 293T cells by transfection with the pcDNA-hRAD51 expression vector. Expression of the heterologous recombinase was checked by western blotting using an anti-FLAG antibody (figure 4A). The global hRAD51 expression level was evaluated by western blotting using an anti-hRAD51 antibody allowing detection of both the expressed and the endogenous hRAD51. Under our experimental conditions, we reached a 3 to 5-fold increase in hRAD51 expression as compared to nontransfected cells or cells transfected with a BAP-FLAG control vector (figure 4B). Immunolocalization experiments of the hRAD51-FLAG protein using an anti-FLAG antibody showed the formation of typical nuclear foci specific of the active DNA repair recombinase while the BAP-FLAG control protein showed a more diffuse localization (figure 4C). This was confirmed by measuring the hRAD51-mediated DNA repair activity through a cisplatin resistance assay, indicating a significant enhancement of the resistance to cisplatin of the cells overexpressing hRAD51-FLAG (figure 4D). Altogether these data demonstrate that hRAD51-FLAG overexpression stimulates the intrinsic cellular DNA repair activity.
Figure 4. Effect of hRAD51 overexpression on endogenous DNA repair and early steps of HIV-1 replication and viral DNA productions.
The expression of hRAD51-FLAG or BAP-FLAG in 293T cells was checked 48 hours post-transfection by western blotting using anti-FLAG antibodies (A, lane 1: protein extract from cells expressing hRAD51-FLAG, lane 2: protein extract from cells expressing BAP-FLAG). The global hRAD51 level was determined in cells transfected with the hRAD51-FLAG (hRAD51) and BAP-FLAG (BAP) expression vectors in parallel to untransfected control cells (w/o transfection), by western blotting using anti-hRAD51 antibodies. The amounts of protein loaded were normalized to the endogenous actin protein revealed by western blotting using an anti-actin antibody (B). The cellular distribution of the overexpressed proteins was determined by immunolocalization using an anti-FLAG antibody (C). The hRAD51 activity was determined under each condition by a cisplatin resistance assay as described in Materials and Method (D). The cells were transduced 48 hours post-transfection with the hRAD51 or BAP expression plasmids. HIV-1 replication was evaluated from fluorescence measurement 10 days after transduction by flow cytometry. The percentage of untransfected eGFP-positive cells was normalized to 100 % (E). The amount of total, integrated and 2-LTR circles viral DNAs was measured by quantitative PCR as described in Materials and Methods. The proportion of the different viral DNA species produced in untransfected control cells was normalized to 100 % (F). Results are represented as the mean values calculated from three independent experiments. The p-values are shown as *p<0.05, **p<0.005.
Early steps of HIV-1 replication were then analyzed in cells overexpressing hRAD51-FLAG by transduction with pRRLsin-PGK-eGFP-WPRE VSV-G pseudotyped viruses and measurement of the eGFP expression from the integrated gene. As reported in figure 4E, hRAD51-FLAG overexpression induced a significant 40–50 % inhibition of HIV-1 replication, in contrast to BAP-FLAG. Under these conditions, no significant change in the total viral DNA amount was detected, while a strong decrease in the integrated DNA forms was observed in addition to an increase of the unintegrated two-LTR circles (figure 4F). This confirmed the effect of hRAD51 on the integration step following nuclear entry of the viral cDNA. To better characterize the relationship between the intracellular concentration of hRAD51 and HIV-1 integration efficiency, we tested whether decreasing the hRAD51 expression level could induce an opposite phenotype. For this purpose, a pharmacological approach was used to reduce hRAD51 expression. Imatinib (figure 5A) was previously reported to decrease the hRAD51 protein levels and increase tumor cell radiosensitivity [17]. In vitro control experiments showed that this compound did not affect HIV-1 IN and hRAD51 catalytic activities (SI9A), validating our approach. As reported in SI9B, treatment with imatinib concentrations above 10 µM led to a significant cellular toxicity (EC50 = 30 ± 8 µM). The effect of the drug on hRAD51 expression was thus evaluated using concentrations below 10 µM. Western blot quantifications of hRAD51 expression following imatinib treatment confirmed that the drug could induce an efficient 40–50% decrease 10 hour post-treatment, while the initial level of hRAD51 was recovered after 72 h (figure 5B). The decrease was also associated with a reduced hRAD51 DNA repair activity as measured by the quantification of cisplatin resistance (figure 5C). Based on these data, HIV-1 replication was tested 24-hour post-treatment. As reported in figure 5D, imatinib treatment 24 hours before transduction of the cells led to an increase in eGFP expression associated with a typical enhancement of the integration efficiency (figure 5E). Taken together, these results demonstrate that an up-regulation of hRAD51 expression both stimulates endogenous DNA repair and induces HIV-1 integration restriction.
Figure 5. Effect of imatinib treatment on hRAD51 expression levels, endogenous DNA repair and early steps of HIV-1 replication.
The chemical structure of imatinib is reported in (A). Total protein fraction was extracted 6 to 72 hours following treatment with imatinib. The hRAD51 protein levels were determined from western blot analyses using an anti-hRAD51 antibody (B). The amount of hRAD51 protein present in untreated cells was normalized to 100%. The ability of imatinib to affect cisplatin resistance was checked in a standard survival analysis performed after 24 hours of treatment with increasing concentrations of the compound (C). Survival was expressed as the ratio of absorbance at 492 nM (Synergy (BioTek) plate reader) of cisplatin-treated cells (pre-incubated with the compound or not) relative to untreated cells. Results represent the means of at least three independent experiments ± standard deviation (error bars).The effect of imatinib on early steps of HIV-1 replication was analyzed following a 24 hours treatment of the cells before transduction with the lentiviral vector. The eGFP fluorescence was measured 10 days post-transduction by flow cytometry. The percentage of eGFP-positive cells in the absence of compound was normalized to 100% (D). The amount of the total, integrated and 2-LTR circles viral DNA forms was quantified at a fixed 10 µM concentration (effective and non-cytotoxic) of imatinib. The amount of each viral DNA species produced in the absence of compound was normalized to 100 % (E). The results are represented as the mean of three independent experiments. The p-values are indicated as *p<0.05, **p<0.005.
hRAD51 expression and intracellular localization are modulated during HIV-1 early steps of replication
To determine whether hRAD51 expression could be modulated during the viral infection we transduced 293T cells with pNL4.3 based pRRLsin-PGK-eGFP-WPRE VSV-G pseudo-typed vectors and analyzed the hRAD51 protein content of cellular extracts obtained at different time points post-transduction. As reported in figure 6A, a decrease in the hRAD51 protein content was detected 4 to 12 hours post-transduction and an increase of the protein level was observed 16–24 hours post-transduction. In contrast, the hRAD51 protein amount was found unchanged in the non-transduced cells. Transcription activity of the RAD51 genes was then analyzed. We used the http://www.peachi.labtelenti.org web resource allowing the querying of cellular responses to infection in SupT1 T cells transduced by HIV-based vectors [18]. Genes encoding for RAD51 and paralogs were found highly downregulated during the early steps of infection (0–6 hours) and upregulated during the integration step (8–16 hours) in contrast to other RAD genes as RAD18 or RAD54 (SI10). Taken together these data indicate that hRAD51 expression is modulated during the early steps of replication at the transcription level. We next analyzed the cellular behavior of the endogenous hRAD51 by immunofluorescence staining of transduced cells at different time points. As shown in figure 6B, a modulation of the cellular localization could also be detected during the early stages of the replication. Indeed while hRAD51 foci were found equally distributed in both cytoplasm and nuclear compartment during the 0 to 8 hours post-transduction a strong relocalization of the recombinase was detected during the 12–24 hours post-transduction. No significant change was observed in the non-transduced cells (S11). Consequently, all these data strongly suggest that viral infection modulates both hRAD51 expression and intracellular localization during the early stages of replication.
Figure 6. Expression (A) and intracellular localization (B) of hRAD51 during the early steps of HIV-1 replication in 293T cells.
Proteins were extracted at different time points (0 to 32 hours) after 293T cells transduction with pNL4.3 based pRRLsin-PGK-eGFP-WPRE VSV-G pseudotyped viruses and the hRAD51 expression level was analyzed by western-blot. The amount of hRAD51 was normalized to the amount of actin detected by western-blot. The quantity of hRAD51 detected at time point = 0 was then normalized to 100 %. The results are represented as the mean of three independent experiments (A). Data obtained without transduction (mock) are also reported. The cellular distribution of hRAD51 was determined by immunolocalization at different time points after 297T cells transduction using an anti-hRAD51 antibody. Results are reported as the mean number ± standard deviation of cytoplasmic and nuclear foci observed under each condition for at least 50 cells (B). A typical picture of hRAD51 intracellular localization is also provided for several time periods corresponding to the early and late steps of integration.
DISCUSSION
Retroviral integration introduces cuts in the host cell DNA that can be considered as a potential mutagenic events by the cellular DNA repair machinery. Furthermore, the incoming intasome containing blunt ended viral DNA can also be recognized as a double strand break by the homologous repair (HR) pathway in the infected cells. This is supported by the formation of active hRAD51 nucleofilaments on viral DNA in presence of IN as detected in previous electron microscopy analyses (11). HR hRAD51 protein binds HIV-1 IN and restricts its activity both in vitro and in vivo through a DNA/IN dissociation process dependent on the formation of active hRAD51 nucleofilaments [6, 7]. The stimulation of hRAD51-mediated inhibition of HIV-1 integration, by using drugs like RS-1, for example, results in the suppression of viral replication in different cell types including primary resting PBMCs [7]. In addition, hRAD51 has been shown to stimulate the expression of proviral genes by enhancing LTR-dependent transcription [10, 11]. Taken together, these data indicate that efficient HIV-1 infection relies on an optimal intracellular activity of hRAD51. Here, using a pharmacological approach allowing to modulate hRAD51 activity, we provide a comprehensive analysis of the mechanisms involved in the regulation of HIV-1 integration.
We first selected molecules capable of modulating the hRAD51-mediated inhibition of HIV-1 IN. Several previously described hRAD51 inhibitors such as RI-1 and A30 [15, 16] were found to alleviate the in vitro integration restriction properties of the recombinase. On the contrary, hRAD51 stimulatory compounds, like the previously reported RS-1 [8] and the newly selected E-pterostilbene purified from grape wine [13], promoted the hRAD51 induced restriction of HIV-1 integration. These results, summarized in SI12, show a strong correlation between the recombination activity of hRAD51 and its ability to inhibit IN. Furthermore none of the drugs affected the IN/hRAD51 association. These data strongly suggest that integration restriction relies on the formation of active hRAD51 nucleofilaments. This nucleocomplex could, thus, constitute a valuable pharmacological target for strategies aiming to modulate HIV-1 replication by targeting the integration step. In addition to provide new information about the hRAD51-induced integration restriction, these molecules could also serve as tools to further explore the role of hRAD51 in proviral LTR driven gene expression.
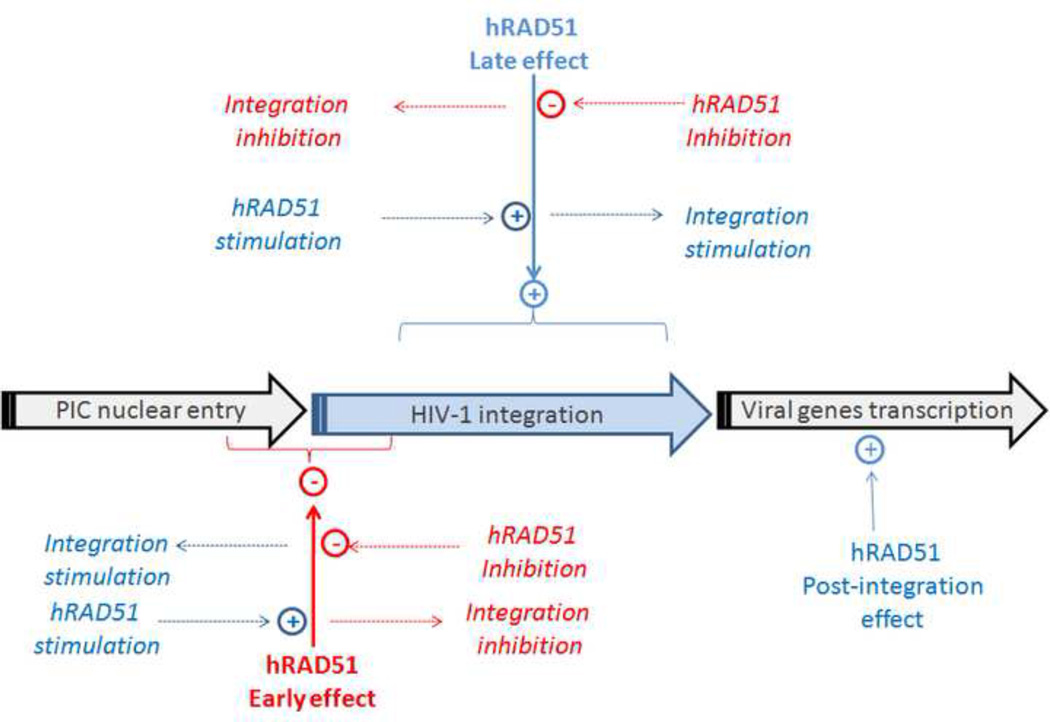
The use of chemical modulators produced differential and opposite effects on both HIV-1 integration and replication depending on the time of treatment (data summarized in SI13). Early stimulation of hRAD51 (treatment at 24-hour pre-transduction) negatively impacted integration and replication whereas late stimulation (i.e. 5-hour post-transduction) had an opposite effect. Likewise, early inhibition of the hRAD51-mediated recombination induced a stimulation of integration and replication, in contrast to what was observed using a 5-hour post-transduction treatment which led to a significant decrease of the replication due to an integration defect. The integration window has been previously determined and is broadly admitted to take place between 6 to 20 hours post-infection in T-cell lines [18, 19]. Since the expression of the reporter gene used in our lentiviral vectors is LTR-independent, our data indicate that hRAD51 plays distinct and opposite functions in the regulation of HIV-1 integration, displaying an early restrictive effect and a late stimulatory effect (summary in figure 7).
Figure 7. Influence of hRAD51 and its modulation on HIV-1 integration.
The hRAD51 recombinase activity, namely the formation of the active nucleofilament, plays a negative regulatory role on the steps following PIC nuclear import and preceding strand transfer catalysis, and a positive regulatory role during the steps subsequent to integration, including DNA repair. Chemical modulators targeting these dual effects can exert opposite effects on HIV-1 integration, and hence on viral replication.
The integration restriction activity of hRAD51 is likely to be related to the previously reported IN/viral cDNA dissociation mechanism through the formation of the hRAD51/DNA nucleofilaments. Indeed, The incoming PICs may be recognized early by hRAD51, thanks to i) the affinity of the recombinase for both IN and the viral cDNA [6, 7], and ii) the recognition of the blunt ended retroviral genome as a double-strand break (DSB), as expected from its intrinsic DNA repair properties [7]. This is also supported by our biochemical data showing that hRAD51 more efficiently targets the HIV-1 IN/DNA complex (see SI1B) and by the systematic correlations observed between HIV-1 integration restriction and hRAD51-mediated DNA repair activity, in vivo (see SI13 and 14). Moreover, under the same experimental conditions, integration inhibition was consistently found associated with an increase of the amount of two-LTR circles assumed to be formed in the nucleus. This indicates an effect downstream of the nuclear import of the PIC. hRAD51 targeting toward the incoming intasome in the nucleus could prevent the latter from reaching the integration locus. This hypothesis is also strongly supported by the treatment-dependent cellular localization of hRAD51, as stimulatory compounds were found to promote the nuclear translocation of the recombinase in contrast to hRAD51 inhibitors (see SI7C–D and SI8B).
The mechanisms of regulation brought into play in cells treated 5 hours post-transduction remain to be fully determined. Nevertheless, the data obtained using stimulatory and inhibitory compounds strongly support that they target a “pro-integrative” function of the recombinase. Since hRAD51 is the main actor of the homologous DNA repair pathway, one relevant hypothesis may be that the recombinase acts at the step of post-integration repair (PIR) by promoting it. This relationship between retroviral integration and DNA repair has been previously proposed [20]. The PIR and the involvement of hRAD51 during this process are currently under study in our laboratory. Especially, the possible role of the nucleofilament-mediated dissociation of HIV-1 IN following strand transfer, which has previously been shown to be required for efficient DNA repair of the integration locus [21, 22], must be investigated. Indeed, this enhancement of the post-integration repair by hRAD51 could be associated to the previously reported function of the recombinase in the LTR-driven transcription of the provirus [9–11] and/or to the chromatin remodeling properties of the recombinase [23]. Indeed, nucleosomal DNA remodeling at the integration site has been proposed to be important for efficient integration [24, 25]. This is supported by the re-localization of endogenous hRAD51 protein during the intregration step in the nuclear compartment where PIR must occur (figure 6B). Biochemical data showing that hRAD51 can promote the in vitro DNA repair step catalyzed by human FEN1 also support this hypothesis (data not shown). However, the role of hRAD51 during the DNA repair of the gapped intermediates remains to be fully investigated and, to this purpose, the chemical compounds reported in this work could constitute useful tools for further studies of this process.
Our data confirm that cellular DNA repair machineries can play multiple roles by restricting retroviral integration and/or directly participating in the stability of the integrated DNA. For an optimal viral replication equilibrium between these opposite effect must be reached. Our results indicate that this can be accomplished by regulation of hRAD51 endogenous expression levels and localization during viral replication (cf figure 6). This suggests that the expression of the anti-integration property of hRAD51 during the early steps of replication (0–6 hours) can be overcome by the decrease of the protein level, and hRAD51 activity. Indeed, this is expected to limit the integration inhibition by the recombinase as observed during chemical treatments with Imatinib decreasing the hRAD51 expression and promoting the integration (figure 5). This would allow the virus to reach a suitable level of efficient integration for optimized replication. In addition to highlight the biological regulatory functions of hRAD51 on HIV-1 integration, our data also provide new insights on the possible use of chemical compounds targeting hRAD51 as antiviral agents restricting HIV-1 replication by modulating the recombinase activity in infected cells. Interestingly the drugs concentrations used in our work are fully compatible with the concentrations used in cellular or clinical studies {Jayathilaka, 2008 #1995; Russell, 2003 #2225}. However, our data indicate that further development towards antiviral compounds will be an hard task due to the dual regulatory function of the recombinase described in this work, as well as the possible secondary effects due to an alteration of DNA repair processes in the infected cells. Nevertheless, we have previously shown that strategies aiming to stimulate hRAD51 activity in a multiple-round infection system, as in PBMCs, could be successful. Indeed, stimulation of the hRAD51-mediated DNA repair pathway prior to HIV-1 infection has shown a protective effect against viral replication without affecting the viability of the cells [7]. Consequently, based on our results, we can speculate that early stimulation of hRAD51 activity (by overexpression or allosteric stimulation) would be efficient against viral replication and effort should be done toward this strategy. Additionally, compounds targeting the hRAD51 DNA repair activity are potential anti-cancer drugs and some anti-cancer molecules, such as imatinib, which is already used in clinic, have an impact on hRAD51 expression. Importantly these effects are observed using micromolar range of drug amounts close to the plasmatic concentrations used in clinic. Consequently, another important conclusion to be drawn from our finding is that great care should be taken when treating HIV-1 infected patients with hRAD51 modulators, as a burst in viral replication may be expected to occur through LTR-dependent proviral transcription and/or under conditions favoring hRAD51 pro-integration functions.
SIGNIFICANCE
Regulation of HIV-1 integration is crucial for efficient viral replication and, thus, constitutes an attractive target for antiviral therapies. The hRAD51 DNA repair protein has been shown to modulate integration activity. A better understanding of the regulation properties of this recombinase is essential for therapeutic approaches targeting DNA repair. The work presented here establishes the link between hRAD51 activity and HIV-1 integration efficiency in infected cells. Indeed, the pharmacological analyses performed demonstrates that hRAD51 plays dual and opposite roles in the regulation of integration, indicating that efficient HIV- replication depends on an optimal intracellular level of hRAD51. In addition to provide new virus/host interactions, our data may have important implications in the development of antiviral strategies based on the modulation of the activity of the cellular DNA repair machinery. Indeed, the work presented here highlights the different pro- and anti-integration properties of hRAD51 that must be taken into consideration for future pharmacological development, and, importantly, shows how chemical modulation of the hRAD51 activity can induce opposite effects on viral replication.
EXPERIMENTAL PROCEDURES
Chemicals, proteins and antibodies
RS-1 and RI-1 were previously selected from a 10,000 compounds library (ChembridgeDIVERSet) screened for their capacity to affect the DNA binding properties of hRAD51 [8],[15]. DIDS previously reported as an hRAD51 inhibitor [12] was purchased from Sigma. E-pterostilbene (P-ter) was extracted from Vitis vinifera leaves (isolation and purification protocols described in [13]). Imatinib was purchased from Selleck chemicals (Euromedex, France). Aptamers A30 and A47, previously selected as ligands of hRAD51 [16], and their shortened versions A30c and A47c were purchased from MWG. Cisplatin was purchased from Sigma. Recombinant HIV-1 IN was expressed in yeast and purified using the INHybrid method previously described [26]. Wild-type recombinant hRAD51 was produced as described before [27]. Monoclonal anti-FLAG and polyclonal anti-hRAD51 antibodies were purchased from Sigma.
In vitro enzymatic assays
HIV-1 concerted integration reactions were performed as described previously [26] and in SI1. All IN activities were quantified by scanning the bands (half-site plus full-site integration products) following gel electrophoresis and autoradiography, using the Image J software. hRAD51 activity was evaluated according to the strand exchange reaction previously reported [28] and adapted to our concerted integration conditions (see SI3). Strand exchange products were quantified following autoradiography of the gel, using the Image J software. Measurements of the hRAD51 DNA binding activity was performed as previously described with slight modifications [15] (see SI2).
Transfection assay
293T cells were grown in DMEM Glutamax® medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 50 µg/ml gentamicin. The cells were seeded into 48-well plates 24 hours before transfection. Fifteen thousand cells were transfected with 1 µg of either p3X-FLAG-CMV-hRAD51 (hRAD51) expression vector or p3X-FLAG-CMV-7-BAP (BAP) expression control plasmid using lipofectamine 2000 (Invitrogen). DMEM containing 20 % of FCS was added 4 hours post-transfection. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 48 hours and then lysed with SDS loading buffer. Extracted proteins were separated on 12 % SDS-PAGE and analyzed by western blotting.
Transduction of 293T cells
293T cells were plated in 48-multiwells plates at 50,000 cells/well using 400 µL of DMEM (Invitrogen, Carlsbad, CA) containing 10% (v/v) fetal calf serum (FCS, Invitrogen) and 50 µg/mL of gentamycin (Invitrogen). Infection was assayed using pNL4.3 based pRRLsin-PGK-eGFP-WPRE VSV-G pseudotyped lentiviruses produced as described in [29]. After three washes with PBS (140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4 pH 7.4), the cells were transduced with the lentiviral vector at an optimized M.O.I of 1, in a final volume of 400 µl (under these conditions, 25–35% of the cells contained one integrated viral cDNA copy). After 10 days, the cells were washed twice with PBS, treated with trypsin for 5 minutes at room temperature and centrifuged 2 minutes at 2000 rpm. The pellet was washed twice with PBS and resuspended in 200 µl PBS/200 mM EDTA. Fluorescence was quantified using 10,000 cells on a FACS Calibur (Beckton-Dickinson, San Jose, CA). Plots were analyzed using the FCS express v3.00.0103. Data are presented as the percentage of cells showing a significant level of fluorescence or as the fluorescence intensity calculated by the X-median fluorescence intensity of all cells.
"V体育ios版" Intracellular hRAD51 immunolocalization
293T cells transfected with the p3X-FLAG-CMV-hRAD51 (hRAD51) or p3X-FLAG-CMV-7-BAP (BAP) expression vectors, or treated with one of our compounds, were washed three times with PBS and incubated for 10 minutes in the fixation buffer containing 2% formaldehyde. After two washes, the cells were incubated 5 minutes in the permeabilization buffer (PBS supplemented with 0.4% saponin and 0.1% Triton X-100). The cells were washed with PBS supplemented with 0.1% saponin and incubated for 1 hour at room temperature in the blocking buffer (PBS 0.1%, 1% BSA, 2% SVF). The cells were incubated with the primary antibody over night at 4 °C. After three washes with PBS, the secondary antibody coupled to Alexa 440 was added and the cells were further incubated 1 hour at 37 °C. DAPI was added at a final concentration of 1 µg/ml for 10 minutes at room temperature. The cells were then washed four times with PBS before fluorescence microscopy analysis. Similar procedure was used for hRAD51 intracellular localization analysis in 293T cells transduced with pRRLsin-PGK-eGFP-WPRE VSV-G pseudotyped lentiviruses.
Cytotoxicity and cell cycle measurements
The cellular cytotoxicity of the compounds was determined by measuring the survival of cells treated with increasing concentrations of molecules for two days with a standard MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay (PromegaCellTiter 96 AQueous One Solution Cell Proliferation Assay). Analyses of the cell cycle were performed using the Propidium iodide flow cytometry kit from abcam following the manufacturer instruction.
Quantification of cisplatin resistance
The effect of protein overexpression and chemical treatment on hRAD51-mediated homologous recombination was assessed using the standard cisplatin resistance assay described previously [8].
Quantification of HIV-1 DNA species
The cells were harvested 8, 24 and 48h post-infection by centrifugation, and 2 to 6.106 cells aliquots were kept frozen at −80°C until analysis. Total DNA (including integrated and episomal HIV-1 DNA) was extracted using the QiAmp blood DNA mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s instructions and eluted in 50 µl elution buffer. Quantification of the viral DNA species was performed using the conditions and primers described in [30]. The total HIV-1 DNA was amplified by quantitative real-time PCR using alight Cycler Instrument (Roche Diagnostics, Meylan, France). Quantifications of total HIV-1 DNA, including 2-LTR circles and integrated HIV-1 cDNA, were performed by quantitative PCR on a Light Cycler instrument (Roche Diagnostics) using the fit point method provided in the Light Cycler quantification software, version 4.1 as previously described [31]. The copy number of 2-LTR circles and total viral DNA were determined with reference to a standard curve prepared by amplification of quantities ranging from 10 to 1.106 copies of plasmid comprising the HIVLAI 2-LTR junction [19]. The integrated HIV-1 cDNA copy number was determined in reference to a standard curve generated by the concomitant two-stage PCR amplification of serial dilutions of an integrated HIV-1 DNA standard from Hela-R7 Neocells [32]. Cell equivalents were calculated according to amplification of the β-globin gene (two copies per diploid cell) with commercially available materials (Control Kit DNA; Roche Diagnostics). 2-LTR circles, total and integrated HIV-1 DNA levels were determined as copy numbers per 106 cells. Two-LTR circles and integrated cDNA were also expressed as a percentage of the total viral DNA.
Supplementary Material
VSports手机版 - Acknowledgments
The work was supported by the French National Research Agency (ANR, RETROSelect program), the French National Research Agency against AIDS (ANRS), SIDACTION, the Centre National de la Recherche Scientifique (CNRS), the University of Bordeaux and the French Research Group GDR 3546. P. Connell and B. Budke were supported by funding from the National Institutes of Health grants (CA142642-02 2010-2015). The authors are deeply grateful to Prof. R. Cooke (University of Bordeaux) for proofreading the manuscript.
Footnotes
AUTHORS CONTRIBUTIONS
TS designed and performed the cellular transduction experiments and viral DNA quantifications, discussed the data, drafted the manuscript; MSB designed and performed the in vitro experiments, discussed the data, drafted the manuscript; LS designed in vitro experiments and discussed the data; ET performed the cellular transduction experiments and viral DNA quantifications; CC purified the recombinant IN proteins; SC carried the mass spectrometry analyses of the IN/hRAD51 interaction (not shown), corrected the manuscript and discussed the data; PWT and JMM purified the stilben compounds and corrected the manuscript; AC performed the hRAD51 transcription study and analyzed the data. PS purified the recombinant hRAD51 protein, discussed the data and corrected the manuscript; PC and BB performed the hRAD51/DNA binding assays, drafted and corrected the manuscript; IH and JH carried secondary experiments in animal models (not shown), discussed the data and corrected the manuscript; MLA discussed the data and corrected the manuscript; OD designed, performed and discussed the viral DNA quantifications, corrected the manuscript; VP set up and coordinated the project, designed and performed the cellular and biochemical experiments, drafted the manuscript and discussed the data.
REFERENCES
- 1.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 2.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A. 107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grandgenett D, Korolev S. Retrovirus Integrase-DNA Structure Elucidates Concerted Integration Mechanisms. Viruses. 2:1185–1189. doi: 10.3390/v2051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherepanov P. Integrase illuminated. EMBO Rep. 11:328. doi: 10.1038/embor.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desfarges S, San Filippo J, Fournier M, Calmels C, Caumont-Sarcos A, Litvak S, Sung P, Parissi V. Chromosomal integration of LTR-flanked DNA in yeast expressing HIV-1 integrase: down regulation by RAD51. Nucleic Acids Res. 2006;34:6215–6224. doi: 10.1093/nar/gkl843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosnefroy O, Tocco A, Lesbats P, Thierry S, Calmels C, Wiktorowicz T, Reigadas S, Kwon Y, De Cian A, Desfarges S, Bonot P, San Filippo J, Litvak S, Cam EL, Rethwilm A, Fleury H, Connell PP, Sung P, Delelis O, Andreola ML, Parissi V. Stimulation of the human RAD51 nucleofilament restricts HIV-1 integration in vitro and in infected cells. J Virol. 2012;86:513–526. doi: 10.1128/JVI.05425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci U S A. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chipitsyna G, Sawaya BE, Khalili K, Amini S. Cooperativity between Rad51 and C/EBP family transcription factors modulates basal and Tat-induced activation of the HIV-1 LTR in astrocytes. J Cell Physiol. 2006;207:605–613. doi: 10.1002/jcp.20612. [DOI] [PubMed] [Google Scholar]
- 10.Kaminski R, Wollebo HS, Datta PK, White MK, Amini S, Khalili K. Interplay of Rad51 with NF-kappaB Pathway Stimulates Expression of HIV-1. PLoS One. 2014;9:e98304. doi: 10.1371/journal.pone.0098304. ["V体育ios版" DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rom I, Darbinyan A, White MK, Rappaport J, Sawaya BE, Amini S, Khalili K. Activation of HIV-1 LTR by Rad51 in microglial cells. Cell Cycle. 2010;9:3715–3722. doi: 10.4161/cc.9.18.12930. [V体育安卓版 - DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida T, Takizawa Y, Kainuma T, Inoue J, Mikawa T, Shibata T, Suzuki H, Tashiro S, Kurumizaka H. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Res. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [V体育安卓版 - DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pflieger A, Waffo Teguo P, Papastamoulis Y, Chaignepain S, Subra F, Munir S, Delelis O, Lesbats P, Calmels C, Andreola ML, Merillon JM, Auge-Gouillou C, Parissi V. Natural Stilbenoids Isolated from Grapevine Exhibiting Inhibitory Effects against HIV-1 Integrase and Eukaryote MOS1 Transposase In Vitro Activities. PLoS One. 2013;8:e81184. doi: 10.1371/journal.pone.0081184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, Connell PP. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. J Med Chem. 2012;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budke B, Logan HL, Kalin JH, Zelivianskaia AS, Cameron McGuire W, Miller LL, Stark JM, Kozikowski AP, Bishop DK, Connell PP. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez SF, Renodon-Corniere A, Nomme J, Eveillard D, Fleury F, Takahashi M, Weigel P. Targeting human Rad51 by specific DNA aptamers induces inhibition of homologous recombination. Biochimie. 2010;92:1832–1838. doi: 10.1016/j.biochi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, Tofilon PJ. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed (VSports手机版)] [Google Scholar]
- 18.Mohammadi P, Desfarges S, Bartha I, Joos B, Zangger N, Munoz M, Gunthard HF, Beerenwinkel N, Telenti A, Ciuffi A. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9:e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manic G, Maurin-Marlin A, Laurent F, Vitale I, Thierry S, Delelis O, Dessen P, Vincendeau M, Leib-Mosch C, Hazan U, Mouscadet JF, Bury-Mone S. Impact of the Ku complex on HIV-1 expression and latency. PLoS One. 2013;8:e69691. doi: 10.1371/journal.pone.0069691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, Bushman FD. Roles of host cell factors in circularization of retroviral dna. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 21.Yoder KE, Bushman FD. Repair of gaps in retroviral DNA integration intermediates. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brin E, Yi J, Skalka AM, Leis J. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J Biol Chem. 2000;275:39287–39295. doi: 10.1074/jbc.M006929200. [DOI] [PubMed] [Google Scholar]
- 23.Dupaigne P, Lavelle C, Justome A, Lafosse S, Mirambeau G, Lipinski M, Pietrement O, Le Cam E. Rad51 polymerization reveals a new chromatin remodeling mechanism. PLoS One. 2008;3:e3643. doi: 10.1371/journal.pone.0003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, Andreola ML, Lavigne M, Parissi V. Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient in vitro Integration into Stable Nucleosomes. PLoS Pathog. 2011;7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesbats P, Lavigne M, Parissi V. HIV-1 integration into chromatin: new insights and future perspective. Future Virology. 2011;6 [VSports - Google Scholar]
- 26.Lesbats P, Metifiot M, Calmels C, Baranova S, Nevinsky G, Andreola ML, Parissi V. In vitro initial attachment of HIV-1 integrase to viral ends: control of the DNA specific interaction by the oligomerization state. Nucleic Acids Res. 2008;36:7043–7058. doi: 10.1093/nar/gkn796. ["VSports手机版" DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Takizawa Y, Kinebuchi T, Kagawa W, Yokoyama S, Shibata T, Kurumizaka H. Mutational analyses of the human Rad51-Tyr315 residue, a site for phosphorylation in leukaemia cells. Genes Cells. 2004;9:781–790. doi: 10.1111/j.1365-2443.2004.00772.x. [VSports手机版 - DOI] [PubMed] [Google Scholar]
- 29.Richard E, Mendez M, Mazurier F, Morel C, Costet P, Xia P, Fontanellas A, Geronimi F, Cario-Andre M, Taine L, Ged C, Malik P, de Verneuil H, Moreau-Gaudry F. Gene therapy of a mouse model of protoporphyria with a self-inactivating erythroid-specific lentiviral vector without preselection. Mol Ther. 2001;4:331–338. doi: 10.1006/mthe.2001.0467. [DOI] [PubMed] [Google Scholar]
- 30.Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamborlini A, Coiffic A, Beauclair G, Delelis O, Paris J, Koh Y, Magne F, Giron ML, Tobaly-Tapiero J, Deprez E, Emiliani S, Engelman A, de The H, Saib A. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J Biol Chem. 286:21013–21022. doi: 10.1074/jbc.M110.189274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munir S, Thierry S, Subra F, Deprez E, Delelis O. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology. 2013;10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
V体育安卓版 - Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.