Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics
- PMID: 24714158
- PMCID: "V体育2025版" PMC3979728
- DOI: 10.1371/journal.pone.0093827
Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics
"V体育平台登录" Abstract
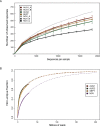
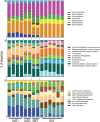
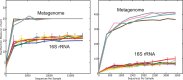
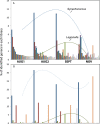
This study explored the short-term planktonic microbial community structure and resilience in Lake Lanier (GA, USA) while simultaneously evaluating the technical aspects of identifying taxa via 16S rRNA gene amplicon and metagenomic sequence data VSports手机版. 16S rRNA gene amplicons generated from four temporally discrete samples were sequenced with 454 GS-FLX-Ti yielding ∼40,000 rRNA gene sequences from each sample and representing ∼300 observed OTUs. Replicates obtained from the same biological sample clustered together but several biases were observed, linked to either the PCR or sequencing-preparation steps. In comparisons with companion whole-community shotgun metagenome datasets, the estimated number of OTUs at each timepoint was concordant, but 1. 5 times and ∼10 times as many phyla and genera, respectively, were identified in the metagenomes. Our analyses showed that the 16S rRNA gene captures broad shifts in community diversity over time, but with limited resolution and lower sensitivity compared to metagenomic data. We also identified OTUs that showed marked shifts in abundance over four close timepoints separated by perturbations and tracked these taxa in the metagenome vs. 16S rRNA amplicon data. A strong summer storm had less of an effect on community composition than did seasonal mixing, which revealed a distinct succession of organisms. This study provides insights into freshwater microbial communities and advances the approaches for assessing community diversity and dynamics in situ. .
Conflict of interest statement
Figures
References
-
- Kent AD, Jones SE, Yannarell AC, Graham JM, Lauster GH, et al. (2004) Annual patterns in bacterioplankton community variability in a humic lake. Microb Ecol 48: 550–560. - VSports注册入口 - PubMed
V体育官网 - Publication types
- "VSports手机版" Actions
MeSH terms
- "VSports在线直播" Actions
- "VSports注册入口" Actions
- Actions (V体育平台登录)
Substances
V体育平台登录 - LinkOut - more resources
Full Text Sources
Other Literature Sources

