V体育官网 - Summary
In T cell-based cancer immunotherapy, tumor antigen (Ag)-specific CD8+ cytotoxic T lymphocytes (CTLs) can specifically target tumor Ags on malignant cells. This promising approach drove us to adopt this strategy of T cell transfer (ACT)-based immunotherapy for chronic viral infections. Here, we describe the generation of hepatitis B virus (HBV) Ag-specific CTLs from induced pluripotent stem cells (iPSCs), i. e. , iPSC-CTLs V体育平台登录. Ag-specific iPSC-CTLs can target HBV Ag+ cells and infiltrate into the liver to suppress HBV replication in a murine model.
For complete details on the use and execution of this protocol, please refer to Haque et al VSports注册入口. (2020).
Subject areas: Immunology, Stem Cells
Graphical Abstract

Highlights (VSports)
-
•
Generation of functional viral Ag-specific CTLs from iPSCs, i V体育官网入口. e. , iPSC-CTLs.
-
•
Viral Ag-specific iPSC-CTLs suppress HBV replication in a murine model
-
•
Adoptive transfer of viral Ag-specific iPSC-CTLs induces persistent α-HBV immunity
-
•
Adoptive transfer of viral Ag-specific iPSC-CTLs produces antiviral cytokines
In T cell-based cancer immunotherapy, tumor antigen (Ag)-specific CD8+ cytotoxic T lymphocytes (CTLs) can specifically target tumor Ags on malignant cells. This promising approach drove us to adopt this strategy of T cell transfer (ACT)-based immunotherapy for chronic viral infections VSports app下载. Here, we describe the generation of hepatitis B virus (HBV) Ag-specific CTLs from induced pluripotent stem cells (iPSCs), i. e. , iPSC-CTLs. Ag-specific iPSC-CTLs can target HBV Ag+ cells and infiltrate into the liver to suppress HBV replication in a murine model.
V体育ios版 - Before you begin
Timing: 0–4 days
-
1.Generation of the retroviral vector and transduction.
-
a.Construct MSCV-IRES-DsRED (MIDR) vector from MSCV-IRES-GFP vector by replacing the GFP gene with the DsRED gene (Lei et al., 2011).
-
b.Generation of HBV construct.
-
i.The HBV 1.2 full length DNA was sub-cloned from the plasmid pHBV-48, containing whole HBV genome to a vector pAAV.
-
ii.A BamHI/EcoRI digested fragment (1.8 kb) and an EcoRI/BglII-digested fragment (2.0 kb) of pHBV-48 were cloned into the BglII site of the AAV vector. The resulting pAAV/HBV1.2 contains the HBV fragments spanning nucleotides 1400-3182.1-1987 flanked by inverted terminal repeats of AAV.
-
i.
-
a.
Note: Prepare all buffers freshly.
-
2.
Retroviral transduction and cell sorting.
Note: Prepare media, reagents, and cell line before the experiments.
-
3.
Platinum-E (Plat-E) packaging cell line is used to generate a pseudo virus for following retroviral transduction.
Note: Prepare all reagents for plasmid preparation.
-
4.
Cells are prepared for cell sorting.
Note: All cell incubations with Gelatin are performed at 37˚C and 230 rpm.
"VSports最新版本" Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 (17A2) | BioLegend | CAT#100236 |
| CD8 (63-6.7) | BioLegend | CAT#100732 |
| CD4 (GK1.5) | BioLegend | CAT#100408 |
| APC IFN-γ (XMG1.2) | BioLegend | CAT#505810 |
| TNF-α (MP6-XT22) | BioLegend | CAT#506306 |
| CD44 (IM7) | BioLegend | CAT#103015 |
| CD69 (H1.2F3) | BioLegend | CAT#104514 |
| CD62L (Mel-14) | BioLegend | CAT#104427 |
| Anti-human TCRα/β | BioLegend | CAT#351711 |
| HRP anti-mouse secondary antibody | Invitrogen | CAT#"V体育2025版" A27025 |
| Anti-HBs antibody | Thermo Fisher | CAT#MA5-13059 |
| Experimental models: cell lines | ||
| Mouse iPS-MEF-Ng-20D-17 | Riken Cell Bank | N/A |
| OP9-DL1/DL4 | ATCC | CRL 2749 |
| SNL 76/7 | ATCC | SCRC-1049 |
| Experimental models: organisms/strains | ||
| HLA-A2.1 transgenic mice | The Pasteur Institute | N/A |
| Recombinant DNA | ||
| HBV construct pAAV/HBV1.2 | Dr. Pei-Jer Chen (National Taiwan University) | N/A |
| Murine hybrid TCR genes Vα34 and Vβ28 | Dr. Adam j. Gehring (Toronto General Hospital Research Institute (TGHRI)) | N/A |
| OVA 257-264 specific TCR genes | Dr. Dario A.A. Vignali (University of Pittsburgh) | N/A |
| BamH1/EcoR1 | New England Biolabs | CAT# R0136T/R0101T |
| EcoRI/BglII | New England Biolabs | CAT# R0101T/R0144M |
| Buffer, media, and reagents | ||
| α-MEM | Invitrogen | CAT# A10490-01 |
| ACK Lysis buffer | Lonza | CAT# 10-548E |
| Brefeldin A | Sigma | CAT# B7651 |
| DMEM | Thermo Fisher | CAT# 11965092 |
| FBS | Hyclone | Product # SH3007.01 |
| Gelatin | Millipore Sigma | CAT# G9391 |
| Gene Jammer | Agilent | CAT# 204130 |
| HLA-A201HBs183-91-PE pentamer | Proimmune | CAT# F027-4A-27 |
| mFlt-3L | Peprotech | CAT# 250-31L |
| mIL-7 | Peprotech | CAT# 217-17 |
| Nuclease S7 | Roche | 10107921001 |
| Paraformaldehyde | Millipore Sigma | SKU P6148-500G |
| Polybrene | Millipore Sigma | SKU 107689 |
| Permeabilization buffer | BioLegend | CAT# 421002 |
| Prolong Gold Antifade Mount with DAPI | Invitrogen | CAT# P36931 |
| QIAamp MinElute Virus Spin Kit | QIAGEN | CAT# 57704 |
| Software and algorithms | ||
| Prism9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Step-by-step method details
VSports手机版 - Cell culture (days 0–5)
-
1.
Murine iPSCs were maintained in DMEM culture medium supplemented with 15% fetal calf serum (FCS), 0. 1 mmol/L nonessential amino acids, 1 mmol/L L-glutamine and 0. 1 mmol/L β-mercaptoethanol. Monolayers of OP9-DL1/DL4 cells were cultured in α-MEM medium supplemented with 20% FCS and 2. 2 g/L sodium bicarbonate VSports最新版本.
-
2.
The iPSCs were washed once in OP9-DL1/DL4 medium before plating onto sub-confluent OP9-DL1/DL4 monolayers for T lineage differentiation in the presence of murine recombinant Flt-3 ligand (mrFlt3L; 5 ng/mL) and murine IL-7 (mIL-7;1 ng/mL).
Generation of HBV viral Ag-specific iPSC-CTLs (days 0–28)
-
3.Mouse iPSCs (iPS-MEF-Ng-20D-17, GFP+) (Yu et al., 2009) were transduced with the MIDR retroviral construct encoding a human-mouse hybrid HBV TCR gene (HBs183-191-specific—s183; TCR Vα34 and Vβ28) (Figure 1A), or OVA257–264 TCR (MiDR-OVA TCR) (Gehring et al., 2011).
-
a.Platinum-E (Plat-E) packaging cell line is used to generate a pseudovirus for following retroviral transduction.
-
b.Seed 3 × 106 Plat-E cells into a 100 mm culture dish containing 10 mL of RPMI complete media with FBS and penicillin/streptomycin 1 day prior to plasmid transfection.
-
c.On day 0, transfect Plat-E cells with HBV TCR-MiDR retroviral vector by using GeneJammer transfection reagent.
-
d.On day 1, take 2 mL of 0.1% Gelatin on to the well of the plate. Keep them in incubator at least for 30 min. After 30 min, remove gelatin from the plate by pipetting. Seed 1 × 106 iPSCs into one well of a 0.1% gelatin-coated 24-well plate containing 2 mL of DMEM media with 15% FBS.
-
e.On day 2, collect pseudovirus-containing supernatant from transfected Plat-E culture and pass the supernatant through a 0.4 μm filter to exclude potential debris and contaminants.
-
f.Perform retroviral transduction in a centrifuge at 500 × g for 1 h in the presence of 5 μg/mL polybrene at 32°C. Use the fresh virus-containing supernatant to get the highest transduction efficiency.
-
g.After transduction, place cells in a 32°C, 5% CO2 incubation overnight (14–16 h).
-
h.On day 3, repeat the second transduction as described above. Meanwhile, pre-coat a 6-well plate with irSNL76/7 feeder cells with α-MEM media at 37˚C for future use.
-
i.On day 4, collect transduced iPSCs by trypsinization, centrifuge at 400 × g for 5 min, and seed into the 6-well plate pre-coated with irSNL76/7 feeder cells with DMEM media supplemented with 15% FBS at 37˚C.
-
j.At confluence, collect cells by trypsinization, centrifuge at 400 × g for 5 min, and process for cell sorting. GFP and DsRED double positive cells will be sorted by a 17-color BD FACS Aria SORP cell sorter. Culture sorted cells (HBV-iPSCs) on fresh irSNL76/7 feeder cells for future use.
-
k.OP9-DL4 and OP9-DL1/DL4 cell lines are generated as described above but by using DL4-MiG and DL1/DL4-MiG constructs, respectively. Transduced cells are sorted by the FACSAria SORP based on GFP expression.
-
a.
-
4.Gene-transduced iPSCs were co-cultured with OP9-DL1/DL4 cells expressing Notch ligands (both DL1 and DL4) molecules in the presence of rFlt3L and rIL-7.
-
a.On day 0, seed 5 × 104 HBV TCR gene-transduced iPSCs into a 100 mm culture dish coated with a confluent monolayer of OP9 (or other OP9-related cell lines) cells in 20% FBS in α-MEM medium at 37˚C.
-
b.On day 3, change culture medium.
-
c.On day 5, collect cells by trypsinization and centrifuge at 400 × g for 5 min before incubating with α-MEM medium on a fresh 100 mm culture dish for 30 min in a 37°C incubator to exclude feeding cells.
-
d.Collect and count floating cells, and transfer 5 × 105 cells to a fresh culture dish containing a confluent monolayer of feeding cells in 20% FBS α-MEM medium. Add the cytokine mFlt-3L (final concentration: 5 ng/mL) into the culture at 37˚C.
-
e.On day 8, gently pipette down loosely attached cells and then wash the feeding layer with 10 mL of PBS to get the maximal recovery of partially differentiated HBV-iPSCs.
-
f.After harvesting cells from the co-culture, centrifuge cells at 400 × g for 5 min and resuspend them in 20 mL of 20% FBS α-MEM medium supplemented with mFlt-3L (5 ng/mL) and IL-7 (1 ng/mL).
-
g.Transfer cells into a 6-well culture plate coated with confluent OP9-feeder cells. Usually HBV TCR-iPSCs recovered from one 100 mm culture dish will be transferred into one well of a 6-well plate.
-
h.From day 10, change medium every other day (20% FBS α-MEM medium supplemented with 5 ng/mL mFlt-3L and 1 ng/mL IL-7).
-
i.Replace OP9 feeder cell plates every 4–6 days depending on the growth of the feeder cells.
-
a.
-
5.
Visualization of the DsRed expression upon gene transduction by fluorescence microscopy (Figure 1 (VSports手机版)B) and sorted GFP+DsRed+ cells (Figure 1 (VSports手机版)C).
-
6.
Confirmation of HBV TCR insertion into the transduced cells by PCR amplification (Figure 1D) and gene sequencing.
-
7.
After 7 days of culture with the OP9-DL1/DL4 cells, iPSCs differentiated into mesoderm-like cells and showed characteristic, non-adherent grape-like cluster morphology on day 14 VSports在线直播. On day 22, lymphocyte-like cells spread fully on the culture plates ("V体育官网" Figure 1E).
-
8.
On day 28 of in vitro co-culture, the iPSC-derived cells substantially expressed CD3 and Ag-specific TCR, the T cell markers.
-
9.
Flow cytometric analysis of CD3+CD8+ populations showed that the HBV s183 but no OVA TCR transduction dramatically increased the generation of HBV-specific CD8+ T cells (CD8+ TCRVβ28+; Figure 1F).
Figure 1.
How to generate HBV viral Ag-specific iPSC-CTLs
Transduce mouse iPSCs with the following retroviral constructs: HBs183-91 TCR (MiDR-HBV s183 TCR) or OVA257–264 TCR (MiDR-OVA TCR), and co-culture the transduced iPSCs with OP9-DL1/DL4 stromal cells for T lineage differentiation.
(A) Present the schematic representation of the retrovirus constructs expressing HBV s183 TCR V体育官网. Ψ, packaging signal; 2A, picornavirus self-cleaving 2A sequence; LTR, long terminal repeats.
(B) Visualize the HBV TCR-transduced iPSCs by a fluorescence microscope.
(C) Transduce GFP+ iPSCs (left and middle, no transduction) with the retroviral construct MiDR or MiDR with HBV s183 TCR, and analyze and sort the GFP+ DsRed+ iPSCs by flow cytometry (right) and by a high-speed cell sorter.
(D) Analyze HBV s183 TCR for Vβ28 gene expression by PCR.
(E) Show morphology of T cell differentiation on various days.
(F) Show flow cytometric analysis of the iPSC-derived cells on day 28, gating on CD3+CD8+ cells, and analyze for the expression of CD8 and TCRVβ28. Data shown are representative of three individual experiments.
Functional assessment of HBV viral Ag-specific iPSC-CTLs (days 28–30)
-
10.
On day 28 of in vitro co-culture, CD4−CD8+ single-positive (SP) iPSC-CTLs were isolated and stimulated them by T-depleted splenocytes pulsed with s183 peptide for indicated time period and assessed cytokine production.
-
11.
The iPSC-CTLs produced large amounts of IL-2 and IFN-γ, as detected by intracellular staining (Figure 2A) or ELISA (Figure 2B) according to the manufacturer instruction and displayed Ag-specific cytotoxicity (Figure 2C), which were similar as HBV TCR gene-transduced CTLs.
Figure 2.
Perform functional analysis of HBV viral Ag-specific iPSC-CTLs
On day 28 of in vitro co-culture, sort the SP CD8+s183 TCR pentamer+ iPSC-T cells. Stimulate the iPSC-T cells and CD8+ T cells transduced with MiDR-s183 TCR with s183 peptide (FLLTRILTI)-pulsed T-depleted splenocytes (APCs) from HHD mice.
(A) Show intracellular staining of IL-2 and IFN-γ after 7 h (gated on CD8+ cells) (T/APCs = 1:4).
(B) Present ELISA of IL-2 and IFN-γ after 40 h.
(C) Measure T cell cytotoxicity after co-culture for 6 h using the 7-AAD/CFSE cell-mediated cytotoxicity assay kit. Data shown are representative of three individual experiments.
Development of HBV replication in mice (weeks 1–8)
-
12.
HLA-A2.1 transgenic (HHD) mice were infected with HBV by hydrodynamic injection of 10 mg of pAAV/HBV1.2 DNA plasmid through the tail veins of mice.
-
13.
Total HBV DNA was resuspended in 1 mL of PBS and delivered into the liver within 3 s. For each animal, 10 μg of pAAV/HBV 1.2 DNA dissolved in 8% body weight of PBS was injected. Before the injection, all animals were anesthetized using isoflurane administered by isoflurane vaporizer. Briefly, anesthetized mice were positioned, so one of the lateral veins are visible. Viral DNA was re suspended in 1 mL of PBS and loaded in 3 ML of sterile syringe with a sterile 27 gauze needle. Position the syringe parallel to the tail with the needle pointing toward the body of the mouse and the bevel of the needle facing up. Proceed with the injection only if the needle is correctly inserted, which is evident from the needle sliding in with no resistance. Press down on plunger in one, continuous motion and inject the full volume of liquid within 3–7 s.
-
14.
Serum level of HBsAg were monitored regularly from the serum of retroorbital blood. The serum was collected from the infected mice on different period of time, serum RNA was collected, and then proceeded for mRNA expression to measure the viral concentration.
-
15.
DNA was isolated from the blood and 100 ng of DNA was used to detect HBV DNA replication using real-time PCR (RT-PCR) followed the manufacturer protocol.
-
16.
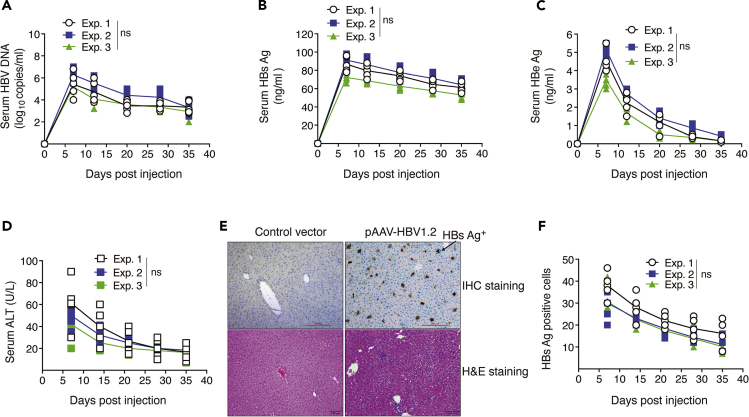
HBV replication were detected in the serum of HHD mice (C57BL/6 background; Figure 3A) from day 3 to 35. DNA replication peaked on day 7 and then reduced gradually. HBV DNA was not cleared from the serum until day 35.
-
17.
We also examined HBs Ag ("VSports在线直播" Figure 3B), HBe Ag ("VSports在线直播" Figure 3C), and alanine aminotransferase (ALT) ("VSports在线直播" Figure 3D) from the serum by ELISA on various days. Similar to DNA replication, HBs Ag peaked on day 7 and then dropped slowly.
-
18.
Intra-hepatic viral DNA transcription and replication were detected from the liver. Mice were sacrificed at different days of post infection, and their livers were isolated and prepared for HBV surface protein staining. In the early period post infection, expression of HBV surface protein was high (approximately 40% of all liver cells are HBs Ag+) and correlates with DNA replication data (VSports - Figures 3E and 3F). During the whole period, we detected HBV protein expression following hydrodynamic injection, although the protein expression was lower than that in the initial period.
-
19.
Inflammatory cell infiltration was detected in the liver after viral infection. Liver samples were processed with hematoxylin and eosin (H&E) staining to detect the inflammatory cells. We observed the robust infiltration of inflammatory cells into the liver in the early period followed by gradually reduction (Figure 3E, lower panel).
Figure 3.
Induce HBV replication in HHD mice by hydrodynamic injection
Perform hydrodynamic tail vein injection of HBV plasmid on HHD mice by injecting 10 μg of plasmid in 8% body weight of PBS.
(A) Show serum HBV copies. On indicated time points after injection, isolate the serum from the blood and extract DNA for RT-PCR.
(B) Present serum HBs Ag.
(C) Show serum HBe Ag.
(D) Show serum ALT. On indicated time points after injection, isolate the serum from the blood and determine protein expression by ELISA.
(E) Present liver tissue histology. Euthanize mice on day 8 after HBV infection. Isolate and stain liver samples for histologic examination.
(F) Show quantitation of HBs Ag+ cells on various days. Data shown are representative of three individual experiments (n = 5).
Detection of accumulated HBV viral Ag-specific iPSC-CTLs in the liver
-
20.
To exert the cytotoxic effects, HBV viral Ag-specific iPSC-CTLs need to accumulate in the liver in which viral replication initiates and expands, and this requires Ag specificity (Tang et al., 2004).
-
21.
To accurately express human HBV TCR on mouse iPSCs, we used murine-human hybrid TCR in which the original human constant region was replaced by that of mouse. Also, for the potential recognition of human TCR in mice, we used HHD mice that express human HLA-A2.1 but lack murine major histocompatibility complex (MHC) class I molecules.
-
22.
Following the hydrodynamic injection of pAAV/HBV1.2 DNA plasmid and adoptive cell transfer of 2 x 106 Ag-specific iPSC-CTLs after 1 week of DNA injection, we used flow cytometry to analyze the expression of HBV-specific TCR on CD8+ T cells. In the spleens and livers of mice receiving HBV-specific iPSC-CTLs, CD8+ T cells expressing HBV- specific TCR were clearly visualized (3.06/11.87 = 25.8% and 1.74/4.5 = 38.7% of CD8+ populations, respectively) but was barely detected in those mice receiving the control cells, i.e., OVA-specific iPSC-CTLs (Figure 4A). Using immunofluorescence staining, we further visualized HBV-specific TCR expression on CD8+T cells in the livers but did not detect its expression on those of mice receiving OVA-specific iPSC- CTLs (Figure 4B).
Figure 4.
Show accumulation of HBV viral Ag-specific iPSC-CTLs in the liver with HBV replication
Euthanize mice at day 8 after the adoptive transfer of T cells. Harvest the spleens or livers and isolate the intra-hepatic lymphocytes by enzymatic digestion.
(A) Analyze accumulation of HBV-specific T cells by flow cytometry. Make single cell suspension from the intra-hepatic lymphocytes and stain cells with fluorochrome-conjugated antibodies. Gate on CD8+ T cells to analyze HBV-specific TCR.
(B) Show immunohistochemistry. Stain liver slides with both fluorochrome-conjugated CD8 antibody and s183 TCR pentamer, and examine under a fluorescent microscope. Green color indicates CD8 and red color indicates HBV-specific TCR. Data are representative of five mice per group of three independent experiments.
"V体育ios版" Expected outcomes
We anticipated that HBV viral Ag-specific iPSC-CTLs will reduce HBV replication in vivo. In order to this, we adoptively transferred HBV viral Ag-specific iPSC-CTLs to detect whether they could prevent or reduce viral replication. Mice were intravenously (i.v.) administrated pAAV/HBV1.2 DNA plasmid via hydrodynamic tail vein injection, and in the following week were i.v. adoptively transferred with HBs183–91 or OVA257–264-specific DsRed+CD8+ iPSC-CTLs. The viral titer was measured by RT-PCR from the serum at various time points after the adoptive transfer of T cells.
The expected outcomes were as followed.
Viral replication was significantly reduced at all time points in the mice receiving HBV viral Ag-specific T cells as compared with the mice receiving the control OVA-specific iPSC-CTLs (Figure 5A).
Figure 5.
Demonstrate in vivo reduction of HBV replication by viral Ag-specific iPSC-CTLs
(A) Show serum HBV copies. At the indicated time points after the adoptive transfer of T cells, isolate the serum from the blood and extract DNA for RT-PCR analysis.
(B) Present liver tissue histology. Euthanize mice on day 8 after the adoptive transfer of T cells. Isolate liver samples and stain for histologic examination. The upper panel shows HBs Ag protein expression (↑) in infected mice (IHC staining) and the lower panel shows the inflammatory cell infiltration (H&E staining).
(C) Present quantitation of HBs Ag positive cells at various days. Data shown are three individual experiments (n = 5).
We also examined the HBV surface protein expression in the livers in the above setting of treatment. Mice were euthanized at various days after the HBV injection, and the liver samples were isolated for histologic examination. Samples were stained for HBV surface protein and examined under a microscope. We observed that HBV surface protein was substantially decreased in the mice receiving HBV viral Ag-specific iPSC-CTLs, as compared with the mice receiving control cells (Figures 5B and 5C), and this is associated with inflammatory cell infiltration visualized by H&E staining.
The mechanism behind the reduction of HBV replication by HBV viral Ag-specific iPSC-CTLs were revealed.
To evaluate the extent to which the antiviral effects reduce HBV replication, we examined the production of the antiviral cytokines IFN-γ and TNF-α from the intra-hepatic lymphocytes of mice receiving HBV viral Ag-specific or OVA-specific iPSC-CTLs. Intra-hepatic lymphocytes were isolated from both groups of mice and stained for IFN-γ and TNF-α. CD8+ T cells were gated, and their productions of IFN-γ and TNF-α were determined by flow cytometry.
The number of IFN-γ-producing CD8+ T cells in the livers was considerably higher in mice receiving HBV viral Ag-specific iPSC-CTLs than in mice receiving the control OVA-specific cells (3.89/22.69 = 17.1% versus <1%; Figures 6A and 6C, upper panel). A similar observation was obtained in the number of TNF-α producing CD8+ T cells (11.85/29.75 = 37.3% versus <1%; Figures 6A and 6C, lower panel).
Figure 6.
Identify the critical role of IFN-γ and TNF-α by viral Ag-specific iPSC-CTLs in the reduction of HBV replication
(A) Show IFN-γ or TNF-α producing CD8+ T cells analyzed by intracellular staining and flow cytometric analysis.
(B) Present immunohistochemistry. Stain liver slides with fluorescence-conjugated CD8 and IFN-γ or TNF-α antibodies, and examine under a fluorescent microscope (red, CD8+; green, IFN-γ+ or TNF-α+).
(C) Show percentage of IFN-γ or TNF-α producing CD8+ T cells.
(D) Show IFN-γ and TNF-α producing CD8+ T cells analyzed by intracellular staining and flow cytometric analysis, gating on CD8+ populations. Data are representative of five mice per group of three independent experiments.
We prepared the liver samples for immunofluorescence staining for detection of IFN-γ or TNF-α-producing CD8+ T cells. More IFN-γ or TNF-α-producing CD8+ T cells were observed in the livers of mice receiving HBV viral Ag-specific iPSC-CTLs than that of mice receiving OVA-specific cells (Figure 6B).
Limitations
Generation of a large number of highly reactive Ag-specific T cells are crucial for adoptive T cell transfer (ACT)-based immunotherapy of various diseases, including cancers (e.g., liver cancer) and autoimmune diseases (e.g., systemic lupus erythematous). Under the appropriate circumstance, iPSCs can produce almost all types of cells in the body, including T cells. Thus, iPSCs provide a chance to obtain a renewable source of healthy T cells for treating a wide array of diseases. However, the optimal circumstance for development of T cells from iPSCs (i.e., iPSC-T cells) has not been fully defined.
The in vitro generation of viral Ag-specific T lymphocytes from stem cells has not been optimized, and the iPSC-derived CD8+ T cells did not differentiate at the same time. In the current study, we developed viral Ag-specific iPSC-CTLs and used them as adoptive cell transfer to reduce HBV replication in a murine model. We sorted DsRed+CD8+ cells for adoptive cell transfer, and the DsRed+CD8+ cells might include CD4+CD8+ (DP) cells and other CD8+ premature lymphocytes. In chronic HBV infection, the viral genome forms a stable mini-chromosome, the covalently closed circular DNA (cccDNA) that can persist throughout the lifespan of the hepatocyte. Targeting the clearance of the viral mini-chromosome may result in a cure of chronic HBV infection. But after the adoptive cell transfer, the premature lymphocytes might change CD8 expression. Nevertheless, the HBV-specific CTLs can be deleted or dysfunctional or succumb to exhaustion in patients with chronic HBV infection (Benechet and Iannacone, 2017, Kawashima et al., 2018). Moreover, recent studies showed that priming by hepatocytes, CD8+ T cells differentiated into dysfunctional HBV-specific CTLs, with partial overlap with those of exhausted or tolerant T cells; thus, these HBV-specific CTLs could not be rescued by treatment with immune checkpoint inhibitors such as anti-PDL1 or CD40-mediated myeloid dendritic cells (mDCs)-activation (Benechet et al., 2019, Isogawa et al., 2013).
Adoptive cell transfer of the HBV-specific CTLs has been considered as a highly promising treatment for chronic HBV infection. The robust mouse models for studying HBV persistence and therapeutic intervention may be the adenovirus and adeno-associated virus (AAV)-based systems, which mimic chronic HBV infection. However, there are still disadvantages in the mouse models in which HBV viruses are transient replication in the liver, and no real HBV viral infection as well as cccDNA formation. We did not detect the obvious destruction of the hepatocytes (Figure 5B), which might be associated with HBV Ag presentation. It has been suggested that, by priming by Kupffer cells, but not hepatocytes, viral Ag-specific CD8+ T cells could efficiently differentiate into functional effector cells against HBV infection (Benechet et al., 2019). In vitro, HBV viral Ag-specific iPSC-CTLs showed the ability to induce specified lysis of target cells, in an effector-to-target ratio-specific manner (Figure 2C), which may not represent in a physiological condition in vivo.
HBV viral Ag-specific iPSC-CTLs characteristically are same as other CTLs. After differentiating into T cells, iPSC-derived cells will not be able to indefinitely proliferate. In fact, iPSC-CTLs have similar growth rate and a confluence as normal T cells. We previously tested the in vivo development of Ag-specific CTLs from iPSCs and did not notice any safety issues of this method (Lei et al., 2011)
Troubleshooting
Problem 1 (cell culture; days 0–5)
Slow growth of mouse iPSCs
Potential solution
Cell culture dish need to be coated properly with sufficient amount of gelatin. Coated dish needs to be in the incubator from 30 min to overnight (14–16 h)
Feeder cells should be confluent in all over the dish but not to be over confluent.
Feeder cells should be healthy and freshly cultured.
Fresh iPSCs should thaw with a minimum temperature and seeded on feeder cells immediately.
iPSCs media should be supplemented with proper supplement and media should be warmed.
iPSCs media should be removed carefully and colony should not be agitated.
Problem 2 (generation of HBV viral Ag-specific iPSC-CTLs; days 0–28)
Inefficient retroviral transduction
Potential solution
Packaging cells should be healthy and freshly prepared.
Transfection reagent should be in optimized concentration.
Centrifuge and incubator should set in appropriate temperature as indicated in methods.
Polybrene concentration should be optimized.
Problem 3 (development of HBV replication in mice; weeks 1–8)
Mice may not be sufficiently infected with HBV
Potential solution
Hydrodynamic delivery of virus is essential for establish HBV mouse model.
Proper syringe and needle should be use.
Large volume of virus suspension should be delivered within 3 s.
Resource availability (V体育ios版)
Lead contact (V体育安卓版)
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianxun Song (jus35@qiuluzeuv.cn).
Materials availability
This study did not generate any unique materials or reagents.
Data and code availability
This study did not generate any unique datasets or code. The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
The authors thank Dr. Pei-Jer Chen from National Taiwan University Hospital for providing the pAAV/HBV1.2 construct and Dr. Adam J. Gehring from Toronto General Hospital Research Institute for providing the s183-specific TCR genes. This work was supported by the National Institutes of Health grants R01AI121180, R01CA221867, and R21AI128325 to J.S.
"V体育平台登录" Author contributions
J.S. designed the experiments, analyzed data, and contributed to the writing of the paper. M.H., F.L., and J.K.D. performed the experiments. X.X. and M.H. analyzed data.
Declaration of interests
The authors declare no competing interests.
References
- Benechet A.P., De Simone G., Di Lucia P., Cilenti F., Barbiera G., Le Bert N., Fumagalli V., Lusito E., Moalli F., Bianchessi V. Dynamics and genomic landscape of CD8(+) T cells undergoing hepatic priming. Nature. 2019;574:200–205. doi: 10.1038/s41586-019-1620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benechet A.P., Iannacone M. Determinants of hepatic effector CD8(+) T cell dynamics. J. Hepatol. 2017;66:228–233. doi: 10.1016/j.jhep.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Gehring A.J., Xue S.A., Ho Z.Z., Teoh D., Ruedl C., Chia A., Koh S., Lim S.G., Maini M.K., Stauss H., Bertoletti A. Engineering virus-specific T cells that target HBV infected hepatocytes and hepatocellular carcinoma cell lines. J. Hepatol. 2011;55:103–110. doi: 10.1016/j.jhep.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Haque M., Lei F., Xiong X., Ren Y., Kumar A., Das J.K., Ren X., Fang D., de Figueiredo P., Yang J.M., Song J. Stem Cell-Derived Viral Antigen-Specific T Cells Suppress HBV Replication through Production of IFN-γ and TNF-⍺. iScience. 2020;23:101333. doi: 10.1016/j.isci.2020.101333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogawa M., Chung J., Murata Y., Kakimi K., Chisari F.V. CD40 activation rescues antiviral CD8(+) T cells from PD-1-mediated exhaustion. PLoS Pathog. 2013;9:e1003490. doi: 10.1371/journal.ppat.1003490. [V体育官网 - DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K., Isogawa M., Hamada-Tsutsumi S., Baudi I., Saito S., Nakajima A., Tanaka Y. Type I interferon signaling prevents hepatitis B virus-specific T cell responses by reducing antigen expression. J. Virol. 2018;92:e01099-18. doi: 10.1128/JVI.01099-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei F., Zhao B., Haque R., Xiong X., Budgeon L., Christensen N.D., Wu Y., Song J. In vivo programming of tumor antigen-specific T lymphocytes from pluripotent stem cells to promote cancer immunosurveillance. Cancer Res. 2011;71:4742–4747. doi: 10.1158/0008-5472.CAN-11-0359. [DOI] [PubMed] [Google Scholar]
- Tang Q., Henriksen K.J., Bi M., Finger E.B., Szot G., Ye J., Masteller E.L., McDevitt H., Bonyhadi M., Bluestone J.A. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 2004;199:1455–1465. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin, Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code. The authors confirm that the data supporting the findings of this study are available within the article.



 Timing: 0–4 days
Timing: 0–4 days