TCF11 Has a Potent Tumor-Repressing Effect Than Its Prototypic Nrf1α by Definition of Both Similar Yet Different Regulatory Profiles, With a Striking Disparity From Nrf2
- PMID: 34268128
- PMCID: PMC8276104 (VSports在线直播)
- DOI: 10.3389/fonc.2021.707032
TCF11 Has a Potent Tumor-Repressing Effect Than Its Prototypic Nrf1α by Definition of Both Similar Yet Different Regulatory Profiles, With a Striking Disparity From Nrf2
Abstract
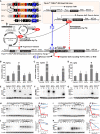
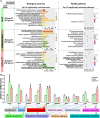
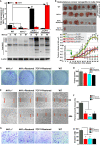
Nrf1 and Nrf2, as two principal CNC-bZIP transcription factors, regulate similar but different targets involved in a variety of biological functions for maintaining cell homeostasis and organ integrity VSports手机版. Of note, the unique topobiological behavior of Nrf1 makes its functions more complicated than Nrf2, because it is allowed for alternatively transcribing and selectively splicing to yield multiple isoforms (e. g. , TCF11, Nrf1α). In order to gain a better understanding of their similarities and differences in distinct regulatory profiles, all four distinct cell models for stably expressing TCF11, TCF11ΔN , Nrf1α or Nrf2 have been herein established by an Flp-In™ T-REx™-293 system and then identified by transcriptomic sequencing. Further analysis revealed that Nrf1α and TCF11 have similar yet different regulatory profiles, although both contribute basically to positive regulation of their co-targets, which are disparate from those regulated by Nrf2. Such disparity in those gene regulations by Nrf1 and Nrf2 was further corroborated by scrutinizing comprehensive functional annotation of their specific and/or common target genes. Conversely, the mutant TCF11ΔN, resulting from a deletion of the N-terminal amino acids 2-156 from TCF11, resembles Nrf2 with the largely consistent structure and function. Interestingly, our further experimental evidence demonstrates that TCF11 acts as a potent tumor-repressor relative to Nrf1α, albeit both isoforms possess a congruous capability to prevent malignant growth of tumor and upregulate those genes critical for improving the survival of patients with hepatocellular carcinoma. .
Keywords: Nrf1α; Nrf2; TCF11; hepatocellular carcinoma; regulatory profiling; transcriptomic sequencing. V体育安卓版.
Copyright © 2021 Wang, Ren, Hu, Liu, Qiu and Zhang V体育ios版. .
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






"V体育ios版" References
LinkOut - more resources (VSports最新版本)
Full Text Sources

