V体育平台登录 - Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma
- PMID: 32686767
- PMCID: PMC7784929
- DOI: V体育官网入口 - 10.1038/s41422-020-0374-x
Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma
Abstract
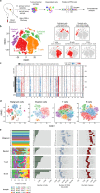
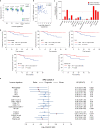
Nasopharyngeal carcinoma (NPC) is an aggressive malignancy with extremely skewed ethnic and geographic distributions. Increasing evidence indicates that targeting the tumor microenvironment (TME) represents a promising therapeutic approach in NPC, highlighting an urgent need to deepen the understanding of the complex NPC TME. Here, we generated single-cell transcriptome profiles for 7581 malignant cells and 40,285 immune cells from fifteen primary NPC tumors and one normal sample. We revealed malignant signatures capturing intratumoral transcriptional heterogeneity and predicting aggressiveness of malignant cells VSports手机版. Diverse immune cell subtypes were identified, including novel subtypes such as CLEC9A+ dendritic cells (DCs). We further revealed transcriptional regulators underlying immune cell diversity, and cell-cell interaction analyses highlighted promising immunotherapeutic targets in NPC. Moreover, we established the immune subtype-specific signatures, and demonstrated that the signatures of macrophages, plasmacytoid dendritic cells (pDCs), CLEC9A+ DCs, natural killer (NK) cells, and plasma cells were significantly associated with improved survival outcomes in NPC. Taken together, our findings represent a unique resource providing in-depth insights into the cellular heterogeneity of NPC TME and highlight potential biomarkers for anticancer treatment and risk stratification, laying a new foundation for precision therapies in NPC. .
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Chen YP, et al. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. - PubMed (V体育平台登录)
-
- Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat. Rev. Cancer. 2016;16:789–802. - VSports手机版 - PubMed
-
- Wang YQ, et al. Prognostic significance of tumor-infiltrating lymphocytes in nondisseminated nasopharyngeal carcinoma: a large-scale cohort study. Int. J. Cancer. 2018;142:2558–2566. - "VSports最新版本" PubMed
-
- Hsu C, et al. Safety and antitumor activity of pembrolizumab in patients with programmed death-ligand 1-positive nasopharyngeal carcinoma: results of the KEYNOTE-028 Study. J. Clin. Oncol. 2017;35:4050–4056. - PubMed
Publication types
MeSH terms
- VSports在线直播 - Actions
- VSports手机版 - Actions
- "V体育2025版" Actions
- "V体育官网" Actions
- "V体育ios版" Actions
- Actions (V体育官网入口)
- "V体育2025版" Actions
- "VSports" Actions
- Actions (VSports在线直播)
- "V体育安卓版" Actions
- VSports手机版 - Actions
"VSports app下载" LinkOut - more resources
Full Text Sources
Molecular Biology Databases

