"V体育平台登录" The LINC00961 transcript and its encoded micropeptide, small regulatory polypeptide of amino acid response, regulate endothelial cell function
- PMID: 31990292
- PMCID: PMC8216332
- DOI: "V体育安卓版" 10.1093/cvr/cvaa008
The LINC00961 transcript and its encoded micropeptide, small regulatory polypeptide of amino acid response, regulate endothelial cell function
Abstract
Aims: Long non-coding RNAs (lncRNAs) play functional roles in physiology and disease, yet understanding of their contribution to endothelial cell (EC) function is incomplete. We identified lncRNAs regulated during EC differentiation and investigated the role of LINC00961 and its encoded micropeptide, small regulatory polypeptide of amino acid response (SPAAR), in EC function. VSports手机版.
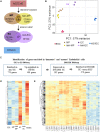
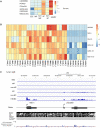
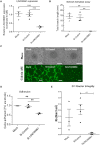
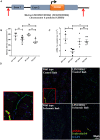
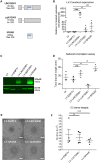
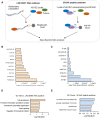
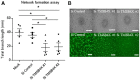
Methods and results: Deep sequencing of human embryonic stem cell differentiation to ECs was combined with Encyclopedia of DNA Elements (ENCODE) RNA-seq data from vascular cells, identifying 278 endothelial enriched genes, including 6 lncRNAs V体育安卓版. Expression of LINC00961, first annotated as an lncRNA but reassigned as a protein-coding gene for the SPAAR micropeptide, was increased during the differentiation and was EC enriched. LINC00961 transcript depletion significantly reduced EC adhesion, tube formation, migration, proliferation, and barrier integrity in primary ECs. Overexpression of the SPAAR open reading frame increased tubule formation; however, overexpression of the full-length transcript did not, despite production of SPAAR. Furthermore, overexpression of an ATG mutant of the full-length transcript reduced network formation, suggesting a bona fide non-coding RNA function of the transcript with opposing effects to SPAAR. As the LINC00961 locus is conserved in mouse, we generated an LINC00961 locus knockout (KO) mouse that underwent hind limb ischaemia (HLI) to investigate the angiogenic role of this locus in vivo. In agreement with in vitro data, KO animals had a reduced capillary density in the ischaemic adductor muscle after 7 days. Finally, to characterize LINC00961 and SPAAR independent functions in ECs, we performed pull-downs of both molecules and identified protein-binding partners. LINC00961 RNA binds the G-actin sequestering protein thymosin beta-4x (Tβ4) and Tβ4 depletion phenocopied the overexpression of the ATG mutant. SPAAR binding partners included the actin-binding protein, SYNE1. .
Conclusion: The LINC00961 locus regulates EC function in vitro and in vivo. The gene produces two molecules with opposing effects on angiogenesis: SPAAR and LINC00961. V体育ios版.
Keywords: Angiogenesis; Endothelial cell; Hind limb ischaemia; LncRNA; Micropeptide VSports最新版本. .
© The Author(s) 2020. Published by Oxford University Press on behalf of the European Society of Cardiology. V体育平台登录.
Figures

V体育ios版 - Comment in
-
V体育官网入口 - Thymosin beta-4x LINCs SPAAR to its non-coding function.Cardiovasc Res. 2020 Oct 1;116(12):1927-1928. doi: 10.1093/cvr/cvaa098. Cardiovasc Res. 2020. PMID: 32282872 Free PMC article. No abstract available.
References
-
- Toborek M, Kaiser S.. Endothelial cell functions. Relationship to atherogenesis. Basic Res Cardiol 1999;94:295–314. - PubMed
-
- De Vries MR, Simons KH, Jukema JW, Braun J, Quax PH.. Vein graft failure: from pathophysiology to clinical outcomes. Nat Rev Cardiol 2016;13:451–470. - PubMed
-
- Pearson J. Normal endothelial cell function. Lupus 2000;9:183–188. - PubMed (VSports在线直播)
Publication types
- Actions (VSports最新版本)
MeSH terms
- VSports最新版本 - Actions
- Actions (VSports在线直播)
- V体育平台登录 - Actions
- Actions (V体育官网入口)
- "V体育平台登录" Actions
- VSports app下载 - Actions
- "VSports最新版本" Actions
- "V体育官网入口" Actions
- Actions (VSports在线直播)
- "V体育官网" Actions
- V体育2025版 - Actions
- "V体育ios版" Actions
- Actions (VSports在线直播)
- Actions (V体育ios版)
- "VSports手机版" Actions
- Actions (VSports最新版本)
- V体育平台登录 - Actions
- "VSports app下载" Actions
- VSports app下载 - Actions
Substances
- "V体育官网" Actions
- "V体育2025版" Actions
- "V体育ios版" Actions
- VSports在线直播 - Actions
- "VSports" Actions
Grants and funding (VSports注册入口)
- RM/17/3/33381/BHF_/British Heart Foundation/United Kingdom (VSports app下载)
- RG/14/3/30706/BHF_/British Heart Foundation/United Kingdom
- FS/16/4/31831/BHF_/British Heart Foundation/United Kingdom
- RM/13/2/30158/BHF_/British Heart Foundation/United Kingdom
- RG/17/4/32662/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

