"V体育官网入口" Linc00961 inhibits the proliferation and invasion of skin melanoma by targeting the miR‑367/PTEN axis
- PMID: 31364744
- PMCID: VSports app下载 - PMC6685588
- DOI: "VSports" 10.3892/ijo.2019.4848
Linc00961 inhibits the proliferation and invasion of skin melanoma by targeting the miR‑367/PTEN axis
Erratum in
-
[Corrigendum] Linc00961 inhibits the proliferation and invasion of skin melanoma by targeting the miR‑367/PTEN axis.Int J Oncol. 2022 Jun;60(6):62. doi: 10.3892/ijo.2022.5352. Epub 2022 Apr 13. Int J Oncol. 2022. PMID: 35417032 Free PMC article.
Abstract
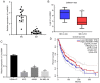
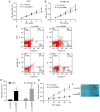
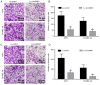
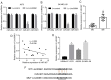
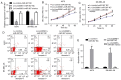
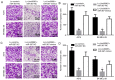
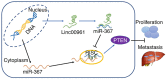
Long intergenic noncoding RNA 00961 (Linc00961) has been identified as a tumor suppressor in various types of cancer. However, the critical roles of Linc00961 in the carcinogenesis and progression of skin melanoma (SM) are yet to be fully elucidated. The present study revealed via reverse transcription‑quantitative PCR analysis that Linc00961 was downregulated in the tissues of patients with SM compared with benign nevi, and in A375, A2058 and SK‑MEL‑28 cell lines compared with human melanocytes. Furthermore, overexpression of Linc00961 inhibited cell proliferation, and promoted the apoptosis of A375 and SK‑MEL‑28 cells in vitro and in vivo, as determined by Cell Counting Kit‑8 and flow cytometry assays, and tumor xenograft studies, respectively. Overexpression of Linc00961 also led to an attenuation of the migration and invasive capabilities of A375 and SK‑MEL‑28 cells, measured using Transwell assays. Functionally, it was demonstrated that Linc00961 acted as a competing endogenous RNA (ceRNA) by competitively sponging microRNA‑367 (miR‑367) in A375 and SK‑MEL‑28 cells; restoration of miR‑367 rescued the inhibitory effects of Linc00961 on A375 and SK‑MEL‑28 cells. Finally, it was observed that phosphate and tension homology deleted on chromosome 10 (PTEN), an established target of miR‑367 in A375 and SK‑MEL‑28 cells, was positively regulated by Linc00961, and its inhibition reversed the inhibitory effects of Linc00961 on the proliferation and invasion of A375 and SK‑MEL‑28 cells VSports手机版. Collectively, the present study revealed that Linc00961 was downregulated in SM, and furthermore, Linc00961 was identified as a ceRNA that inhibits the proliferation and invasion of A375 and SK‑MEL‑28 cells by modulating the miR‑367/PTEN axis. .
Figures



References
-
- Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. - DOI (VSports注册入口) - PMC - PubMed
V体育ios版 - MeSH terms
- Actions (VSports)
- "V体育官网" Actions
- V体育官网入口 - Actions
- "V体育安卓版" Actions
- "V体育官网" Actions
- "V体育官网入口" Actions
- VSports最新版本 - Actions
- Actions (VSports最新版本)
- V体育官网入口 - Actions
- "VSports" Actions
- "VSports在线直播" Actions
- "V体育平台登录" Actions
- V体育官网入口 - Actions
Substances
- "VSports在线直播" Actions
- VSports app下载 - Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

