High mobility group box 1 enables bacterial lipids to trigger receptor-interacting protein kinase 3 (RIPK3)-mediated necroptosis and apoptosis in mice
- PMID: 31000631
- PMCID: VSports在线直播 - PMC6552418
- DOI: 10.1074/jbc.RA118.007040 (V体育平台登录)
High mobility group box 1 enables bacterial lipids to trigger receptor-interacting protein kinase 3 (RIPK3)-mediated necroptosis and apoptosis in mice
Erratum in
-
Correction: High mobility group box 1 enables bacterial lipids to trigger receptor-interacting protein kinase 3 (RIPK3)-mediated necroptosis and apoptosis in mice.J Biol Chem. 2019 Aug 9;294(32):12261. doi: 10.1074/jbc.AAC119.010124. J Biol Chem. 2019. PMID: 31399536 Free PMC article. No abstract available.
Abstract
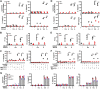
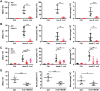
Receptor-interacting protein kinase 3 (RIPK3) is a key regulator of programmed cell death and inflammation during viral infection or sterile tissue injury. Whether and how bacterial infection also activates RIPK3-dependent immune responses remains poorly understood. Here we show that bacterial lipids (lipid IVa or lipid A) form a complex with high mobility group box 1 (HMGB1), released by activated immune cells or damaged tissue during bacterial infection, and that this complex triggers RIPK3- and TIR domain-containing adapter-inducing IFN-β (TRIF)-dependent immune responses. We found that these responses lead to macrophage death, interleukin (IL)-1α release, and IL-1β maturation. In an air-pouch inflammatory infiltration model, genetic deletion of Ripk3, Trif, or IL-1 receptor (Il-1R), or monoclonal antibody-mediated HMGB1 neutralization uniformly attenuated inflammatory responses induced by Gram-negative bacteria that release lipid IVa and lipid A. These findings uncover a previously unrecognized mechanism by which host factors and bacterial components work in concert to orchestrate immune responses VSports手机版. .
Keywords: HMGB1; RIPK3; apoptosis; bacteria; damage-associated molecular patterns; inflammasome; lipid A; necroptosis; tumor necrosis factor (TNF) V体育安卓版. .
© 2019 Meng et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
"VSports app下载" Figures





References
-
- Lu B., Wang C., Wang M., Li W., Chen F., Tracey K. J., and Wang H. (2014) Molecular mechanism and therapeutic modulation of high mobility group box 1 release and action: an updated review. Expert Rev. Clin. Immunol. 10, 713–727 10.1586/1744666X.2014.909730 - "V体育官网" DOI - PMC - PubMed
-
- Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundbäck P., Valdes-Ferrer S. I., Olofsson P. S., Kalb T., Roth J., Zou Y., Erlandsson-Harris H., Yang H., Ting J. P., Wang H. et al. (2012) Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670–674 10.1038/nature11290 - DOI - PMC - PubMed
-
- Lu B., Antoine D. J., Kwan K., Lundbäck P., Wähämaa H., Schierbeck H., Robinson M., Van Zoelen M. A., Yang H., Li J., Erlandsson-Harris H., Chavan S. S., Wang H., et al. (2014) JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc. Natl. Acad. Sci. U.S.A. 111, 3068–3073 10.1073/pnas.1316925111 - DOI - PMC - PubMed
Publication types
MeSH terms (V体育ios版)
- VSports手机版 - Actions
- "V体育安卓版" Actions
- "VSports app下载" Actions
- Actions (VSports手机版)
- "V体育ios版" Actions
- "V体育官网" Actions
- Actions (VSports手机版)
- "VSports注册入口" Actions
- "V体育官网入口" Actions
- V体育ios版 - Actions
- V体育平台登录 - Actions
Substances
- V体育官网 - Actions
- Actions (V体育平台登录)
- Actions (VSports手机版)
- "VSports注册入口" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

