Nrf2 promotes breast cancer cell migration via up-regulation of G6PD/HIF-1α/Notch1 axis
- PMID: 30809937
- PMCID: PMC6484400
- DOI: 10.1111/jcmm.14241
Nrf2 promotes breast cancer cell migration via up-regulation of G6PD/HIF-1α/Notch1 axis
"V体育ios版" Abstract
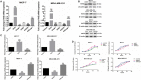
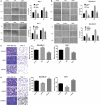
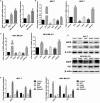
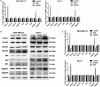
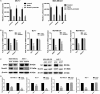
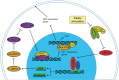
Abnormal metabolism of tumour cells is closely related to the occurrence and development of breast cancer, during which the expression of NF-E2-related factor 2 (Nrf2) is of great significance. Metastatic breast cancer is one of the most common causes of cancer death worldwide; however, the molecular mechanism underlying breast cancer metastasis remains unknown VSports手机版. In this study, we found that the overexpression of Nrf2 promoted proliferation and migration of breast cancers cells. Inhibition of Nrf2 and overexpression of Kelch-like ECH-associated protein 1 (Keap1) reduced the expression of glucose-6-phosphate dehydrogenase (G6PD) and transketolase of pentose phosphate pathway, and overexpression of Nrf2 and knockdown of Keap1 had opposite effects. Our results further showed that the overexpression of Nrf2 promoted the expression of G6PD and Hypoxia-inducing factor 1α (HIF-1α) in MCF-7 and MDA-MB-231 cells. Overexpression of Nrf2 up-regulated the expression of Notch1 via G6PD/HIF-1α pathway. Notch signalling pathway affected the proliferation of breast cancer by affecting its downstream gene HES-1, and regulated the migration of breast cancer cells by affecting the expression of EMT pathway. The results suggest that Nrf2 is a potential molecular target for the treatment of breast cancer and targeting Notch1 signalling pathway may provide a promising strategy for the treatment of Nrf2-driven breast cancer metastasis. .
Keywords: G6PD; HIF-1α; Notch1; Nrf2; breast cancer V体育安卓版. .
© 2019 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine. V体育ios版.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Hu ZI, McArthur HL. Immunotherapy in breast cancer: the New Frontier. Curr Breast Cancer Rep. 2018;10:35‐40. - PMC (VSports最新版本) - PubMed
-
- Walker A, Singh A, Tully E, et al. Nrf2 signaling and autophagy are complementary in protecting breast cancer cells during glucose deprivation. Free Radic Biol Med. 2018;120:407‐413. - "V体育2025版" PMC - PubMed
VSports app下载 - Publication types
MeSH terms
- Actions (VSports在线直播)
- V体育安卓版 - Actions
- Actions (V体育安卓版)
- V体育2025版 - Actions
- Actions (V体育ios版)
- "V体育平台登录" Actions
- Actions (VSports注册入口)
- "VSports在线直播" Actions
Substances
- V体育官网入口 - Actions
- VSports在线直播 - Actions
- V体育官网 - Actions
Associated data
- VSports手机版 - Actions
- VSports手机版 - Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports在线直播 - Medical
Research Materials
Miscellaneous

