Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation
- PMID: 29447696
- PMCID: V体育官网入口 - PMC8318347
- DOI: 10.1016/j.chom.2018.01.003
Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation
Abstract
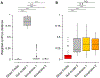
Fecal microbiota transplantation (FMT) from healthy donor to patient is a treatment for microbiome-associated diseases. Although the success of FMT requires donor bacteria to engraft in the patient's gut, the forces governing engraftment in humans are unknown. Here we use an ongoing clinical experiment, the treatment of recurrent Clostridium difficile infection, to uncover the rules of engraftment in humans. We built a statistical model that predicts which bacterial species will engraft in a given host, and developed Strain Finder, a method to infer strain genotypes and track them over time. We find that engraftment can be predicted largely from the abundance and phylogeny of bacteria in the donor and the pre-FMT patient. Furthermore, donor strains within a species engraft in an all-or-nothing manner and previously undetected strains frequently colonize patients receiving FMT VSports手机版. We validated these findings for metabolic syndrome, suggesting that the same principles of engraftment extend to other indications. .
Keywords: C V体育安卓版. difficile; Clostridium difficile; FMT; bacterial engraftment; fecal microbiota transplant; fecal transplant; human microbiome; human microbiota; strain inference; strain tracking. .
Copyright © 2018 Elsevier Inc. All rights reserved. V体育ios版.
Figures

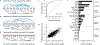


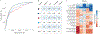

Comment in (V体育2025版)
-
Therapy: FMT: the rules of engraftment.Nat Rev Gastroenterol Hepatol. 2018 Apr;15(4):190-191. doi: 10.1038/nrgastro.2018.19. Epub 2018 Feb 28. Nat Rev Gastroenterol Hepatol. 2018. PMID: 29487424 No abstract available.
V体育2025版 - References
-
- Aroniadis OC, and Brandt LJ (2013). Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol 29, 79–84. - PubMed (V体育官网入口)
-
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. - PubMed
-
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, and Young VB (2008). Decreased Diversity of the Fecal Microbiome in Recurrent Clostridium difficile—Associated Diarrhea. J. Infect. Dis 197, 435–438. - PubMed (V体育官网)
MeSH terms
- V体育2025版 - Actions
- "VSports在线直播" Actions
- Actions (V体育官网)
- "V体育安卓版" Actions
- Actions (V体育ios版)
Grants and funding (VSports注册入口)
LinkOut - more resources
Full Text Sources
"VSports" Other Literature Sources

