"V体育官网" A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients
- PMID: 28174747
- PMCID: PMC5247316
- DOI: "V体育官网入口" 10.1016/j.jcmgh.2016.06.004
A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients
"VSports在线直播" Abstract
Background & aims: Microbes may increase susceptibility to inflammatory bowel disease (IBD) by producing bioactive metabolites that affect immune activity and epithelial function VSports手机版. We undertook a family based study to identify microbial and metabolic features of IBD that may represent a predisease risk state when found in healthy first-degree relatives. .
Methods: Twenty-one families with pediatric IBD were recruited, comprising 26 Crohn's disease patients in clinical remission, 10 ulcerative colitis patients in clinical remission, and 54 healthy siblings/parents. Fecal samples were collected for 16S ribosomal RNA gene sequencing, untargeted liquid chromatography-mass spectrometry metabolomics, and calprotectin measurement. Individuals were grouped into microbial and metabolomics states using Dirichlet multinomial models. Multivariate models were used to identify microbes and metabolites associated with these states. V体育安卓版.
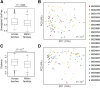
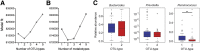
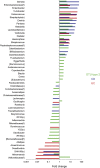
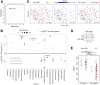
Results: Individuals were classified into 2 microbial community types. One was associated with IBD but irrespective of disease status, had lower microbial diversity, and characteristic shifts in microbial composition including increased Enterobacteriaceae, consistent with dysbiosis. This microbial community type was associated similarly with IBD and reduced microbial diversity in an independent pediatric cohort. Individuals also clustered bioinformatically into 2 subsets with shared fecal metabolomics signatures. One metabotype was associated with IBD and was characterized by increased bile acids, taurine, and tryptophan. The IBD-associated microbial and metabolomics states were highly correlated, suggesting that they represented an integrated ecosystem. Healthy relatives with the IBD-associated microbial community type had an increased incidence of elevated fecal calprotectin. V体育ios版.
Conclusions: Healthy first-degree relatives can have dysbiosis associated with an altered intestinal metabolome that may signify a predisease microbial susceptibility state or subclinical inflammation VSports最新版本. Longitudinal prospective studies are required to determine whether these individuals have a clinically significant increased risk for developing IBD. .
Keywords: AUC, area under the curve; CD, Crohn’s disease; Family Cohort; IBD, inflammatory bowel disease; Inflammatory Bowel Disease; LC/MS, liquid chromatography/mass spectrometry; Metabolomics; Microbiome; OTU, operational taxonomic unit; PCR, polymerase chain reaction; PCoA, principal coordinates analysis; ToFMS, time-of-flight mass spectrometry; UC, ulcerative colitis; UPLC, ultra-performance liquid chromatography; rRNA, ribosomal RNA. V体育平台登录.
Figures




References
-
- Ng S.C., Bernstein C.N., Vatn M.H. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630–649. - "VSports最新版本" PubMed
-
- McGovern D.P., Kugathasan S., Cho J.H. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149:1163–1176 e2. - "V体育平台登录" PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

