V体育官网 - A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor
- PMID: 27909575
- PMCID: "VSports app下载" PMC5112579
- DOI: 10.12688/f1000research.9501.2
"VSports手机版" A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor
VSports在线直播 - Abstract
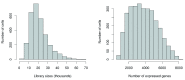
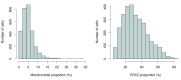
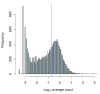
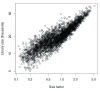
Single-cell RNA sequencing (scRNA-seq) is widely used to profile the transcriptome of individual cells. This provides biological resolution that cannot be matched by bulk RNA sequencing, at the cost of increased technical noise and data complexity. The differences between scRNA-seq and bulk RNA-seq data mean that the analysis of the former cannot be performed by recycling bioinformatics pipelines for the latter VSports手机版. Rather, dedicated single-cell methods are required at various steps to exploit the cellular resolution while accounting for technical noise. This article describes a computational workflow for low-level analyses of scRNA-seq data, based primarily on software packages from the open-source Bioconductor project. It covers basic steps including quality control, data exploration and normalization, as well as more complex procedures such as cell cycle phase assignment, identification of highly variable and correlated genes, clustering into subpopulations and marker gene detection. Analyses were demonstrated on gene-level count data from several publicly available datasets involving haematopoietic stem cells, brain-derived cells, T-helper cells and mouse embryonic stem cells. This will provide a range of usage scenarios from which readers can construct their own analysis pipelines. .
Keywords: Bioconductor; RNA-seq; Single cell; bioinformatics; workflow V体育安卓版. .
Conflict of interest statement (VSports在线直播)
Figures





















References
-
- Anders S, Huber W: Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. 10.1186/gb-2010-11-10-r106 - V体育官网入口 - DOI - PMC - PubMed
-
- Bourgon R, Gentleman R, Huber W: Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107(21):9546–9551. 10.1073/pnas.0914005107 - "VSports app下载" DOI - PMC - PubMed
"VSports" Grants and funding
LinkOut - more resources
"VSports在线直播" Full Text Sources
"VSports app下载" Other Literature Sources
Molecular Biology Databases

