"VSports" Consequences of bile salt biotransformations by intestinal bacteria
- PMID: 26939849
- PMCID: VSports注册入口 - PMC4856454
- DOI: 10.1080/19490976.2015.1127483
"V体育官网" Consequences of bile salt biotransformations by intestinal bacteria
"V体育ios版" Erratum in
-
Erratum: Publisher's note.Gut Microbes. 2016 Jun 9;7(3):262. doi: 10.1080/19490976.2016.1191920. eCollection 2016. Gut Microbes. 2016. PMID: 31265699 Free PMC article.
Abstract
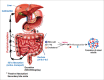
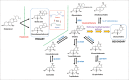
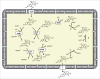
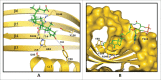
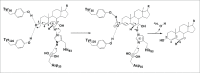
Emerging evidence strongly suggest that the human "microbiome" plays an important role in both health and disease. Bile acids function both as detergents molecules promoting nutrient absorption in the intestines and as hormones regulating nutrient metabolism. Bile acids regulate metabolism via activation of specific nuclear receptors (NR) and G-protein coupled receptors (GPCRs) VSports手机版. The circulating bile acid pool composition consists of primary bile acids produced from cholesterol in the liver, and secondary bile acids formed by specific gut bacteria. The various biotransformation of bile acids carried out by gut bacteria appear to regulate the structure of the gut microbiome and host physiology. Increased levels of secondary bile acids are associated with specific diseases of the GI system. Elucidating methods to control the gut microbiome and bile acid pool composition in humans may lead to a reduction in some of the major diseases of the liver, gall bladder and colon. .
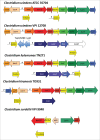
Keywords: bile acid 7α-dehydroxylation; bile acid oxidoreductases; bile salt hydrolase; gut microbiota; secondary bile acids. V体育安卓版.
"V体育平台登录" Figures

References
-
- Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology 1978; 75(1):71–5; PMID:401099 - PubMed
-
- Dawaon PA, Karpen SJ. J Lipid Res 2015; 56(6):1085–99; PMID:25210150; VSports app下载 - http://dx.doi.org/ 10.1194/jlr.R054114 - DOI - PMC - PubMed
-
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science 1999; 284:1362–5; PMID:10334992; VSports注册入口 - http://dx.doi.org/ 10.1126/science.284.5418.1362 - DOI - PubMed
-
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Wilson TM, Zavacki AM, Moore DD, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science 1999; 284:1365-8. - PubMed
-
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y et al. A G-protein-coupled receptor responsive to bile acids. J Biol Chem 2003; 278:9435–40 - PubMed
Publication types
MeSH terms
- VSports最新版本 - Actions
- V体育官网入口 - Actions
- VSports在线直播 - Actions
Substances
- V体育安卓版 - Actions
- "V体育2025版" Actions
- VSports最新版本 - Actions
V体育官网入口 - Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育ios版" Molecular Biology Databases
