V体育官网入口 - Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut
- PMID: 26506949
- PMCID: PMC4623478
- DOI: 10.1038/srep15782
Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut
Abstract
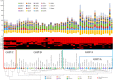
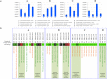
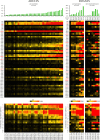
Bifidobacteria are common and frequently dominant members of the gut microbiota of many animals, including mammals and insects. Carbohydrates are considered key carbon sources for the gut microbiota, imposing strong selective pressure on the complex microbial consortium of the gut VSports手机版. Despite its importance, the genetic traits that facilitate carbohydrate utilization by gut microbiota members are still poorly characterized. Here, genome analyses of 47 representative Bifidobacterium (sub)species revealed the genes predicted to be required for the degradation and internalization of a wide range of carbohydrates, outnumbering those found in many other gut microbiota members. The glycan-degrading abilities of bifidobacteria are believed to reflect available carbon sources in the mammalian gut. Furthermore, transcriptome profiling of bifidobacterial genomes supported the involvement of various chromosomal loci in glycan metabolism. The widespread occurrence of bifidobacterial saccharolytic features is in line with metagenomic and metatranscriptomic datasets obtained from human adult/infant faecal samples, thereby supporting the notion that bifidobacteria expand the human glycobiome. This study also underscores the hypothesis of saccharidic resource sharing among bifidobacteria through species-specific metabolic specialization and cross feeding, thereby forging trophic relationships between members of the gut microbiota. .
"VSports手机版" Figures

References
-
- Ventura M., Turroni F., Motherway M. O., MacSharry J. & van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends in microbiology 20, 467–476, doi: 10.1016/j.tim.2012.07.002 (2012). - "V体育平台登录" DOI - PubMed
-
- Bergey D. H., Goodfellow M., Whitman W. B. & Parte A. C. 1 online resource (2 v.) (Springer, New York, 2012).
-
- Turroni F. et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Applied and environmental microbiology 75, 1534–1545, doi: 10.1128/AEM.02216-08 (2009). - "V体育安卓版" DOI - PMC - PubMed
"V体育平台登录" Publication types
MeSH terms
- "VSports" Actions
- "VSports手机版" Actions
- "V体育官网" Actions
Substances
"VSports在线直播" Grants and funding
LinkOut - more resources
"V体育平台登录" Full Text Sources
Other Literature Sources
VSports手机版 - Medical

