VSports最新版本 - Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates
- PMID: 26184873
- PMCID: PMC4666378
- DOI: VSports app下载 - 10.1093/nar/gkv715
Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates
Abstract
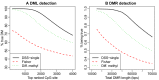
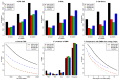
DNA methylation is an important epigenetic modification involved in many biological processes and diseases. Recent developments in whole genome bisulfite sequencing (WGBS) technology have enabled genome-wide measurements of DNA methylation at single base pair resolution. Many experiments have been conducted to compare DNA methylation profiles under different biological contexts, with the goal of identifying differentially methylated regions (DMRs). Due to the high cost of WGBS experiments, many studies are still conducted without biological replicates. Methods and tools available for analyzing such data are very limited. We develop a statistical method, DSS-single, for detecting DMRs from WGBS data without replicates. We characterize the count data using a rigorous model that accounts for the spatial correlation of methylation levels, sequence depth and biological variation. We demonstrate that using information from neighboring CG sites, biological variation can be estimated accurately even without replicates. DMR detection is then carried out via a Wald test procedure. Simulations demonstrate that DSS-single has greater sensitivity and accuracy than existing methods, and an analysis of H1 versus IMR90 cell lines suggests that it also yields the most biologically meaningful results. DSS-single is implemented in the Bioconductor package DSS. VSports手机版.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research V体育安卓版. .
"VSports在线直播" Figures
V体育官网入口 - References
-
- Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Scholer A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D., et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. - PubMed
-
- Hansen K.D., Timp W., Bravo H.C., Sabunciyan S., Langmead B., McDonald O.G., Wen B., Wu H., Liu Y., Diep D., et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43:768–775. - VSports app下载 - PMC - PubMed
-
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
-
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. - PubMed
MeSH terms
- V体育安卓版 - Actions
- Actions (V体育2025版)
- "V体育官网" Actions
Substances
- "V体育官网" Actions
"V体育官网入口" Grants and funding
LinkOut - more resources
Full Text Sources
"V体育平台登录" Other Literature Sources

