Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes
- PMID: 25517115
- PMCID: VSports app下载 - PMC4269443
- DOI: 10.1371/journal.pone.0115175
Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes (V体育2025版)
Abstract
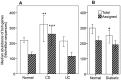
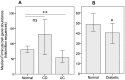
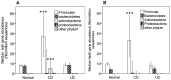
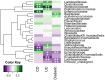
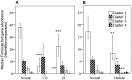
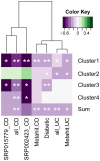
We performed an analysis to determine the importance of bile acid modification genes in the gut microbiome of inflammatory bowel disease and type 2 diabetic patients. We used publicly available metagenomic datasets from the Human Microbiome Project and the MetaHIT consortium, and determined the abundance of bile salt hydrolase gene (bsh), 7 alpha-dehydroxylase gene (adh) and 7-alpha hydroxysteroid dehydrogenase gene (hsdh) in fecal bacteria in diseased populations of Crohn's disease (CD), Ulcerative Colitis (UC) and Type 2 diabetes mellitus (T2DM). Phylum level abundance analysis showed a significant reduction in Firmicute-derived bsh in UC and T2DM patients but not in CD patients, relative to healthy controls. Reduction of adh and hsdh genes was also seen in UC and T2DM patients, while an increase was observed in the CD population as compared to healthy controls. A further analysis of the bsh genes showed significant differences in the correlations of certain Firmicutes families with disease or healthy populations. From this observation we proceeded to analyse BSH protein sequences and identified BSH proteins clusters representing the most abundant strains in our analysis of Firmicute bsh genes. The abundance of the bsh genes corresponding to one of these protein clusters was significantly reduced in all disease states relative to healthy controls. This cluster includes bsh genes derived from Lachospiraceae, Clostridiaceae, Erysipelotrichaceae and Ruminococcaceae families. This metagenomic analysis provides evidence of the importance of bile acid modifying enzymes in health and disease. It further highlights the importance of identifying gene and protein clusters, as the same gene may be associated with health or disease, depending on the strains expressing the enzyme, and differences in the enzymes themselves. VSports手机版.
"V体育ios版" Conflict of interest statement
Competing Interests: Micropharma Limited funded this research. MLJ and SP are co-founders and shareholders of Micropharma. MLJ, CJM, JGG and AL are employed by and CJM is a shareholder of Micropharma. AL, JGG and MLJ designed the research. AL collected and analyzed the data. AL, JGG, CJM and MLJ prepared the manuscript for publication V体育安卓版. Micropharma Limited holds no editorial restriction over the work published and the authors' association with it does not alter their adherence to PLOS ONE policies on sharing data and materials.
Figures
"V体育安卓版" References
-
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500 UR: 541–546. - PubMed
-
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R, et al. (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489 UR: 220–230. - PMC (VSports手机版) - PubMed
-
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B, et al. (2009) Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 89 UR: 147–191. - PubMed
"VSports app下载" Publication types
MeSH terms
- "VSports在线直播" Actions
- "VSports手机版" Actions
- V体育安卓版 - Actions
- Actions (VSports最新版本)
- "V体育平台登录" Actions
- V体育安卓版 - Actions
- V体育安卓版 - Actions
- Actions (VSports最新版本)
- "VSports" Actions
- "VSports" Actions
- Actions (VSports app下载)
- VSports注册入口 - Actions
- VSports最新版本 - Actions
Substances
- "V体育官网入口" Actions
- Actions (V体育ios版)
- "VSports手机版" Actions
"V体育平台登录" LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources
Medical

