"VSports app下载" Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs
- PMID: 25071214
- PMCID: VSports注册入口 - PMC4143054
- DOI: "VSports最新版本" 10.1073/pnas.1410622111
Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs
V体育官网入口 - Abstract
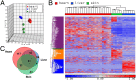
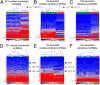
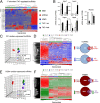
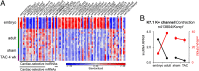
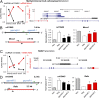
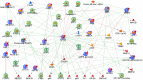
The vast majority of mammalian DNA does not encode for proteins but instead is transcribed into noncoding (nc)RNAs having diverse regulatory functions. The poorly characterized subclass of long ncRNAs (lncRNAs) can epigenetically regulate protein-coding genes by interacting locally in cis or distally in trans. A few reports have implicated specific lncRNAs in cardiac development or failure, but precise details of lncRNAs expressed in hearts and how their expression may be altered during embryonic heart development or by adult heart disease is unknown. Using comprehensive quantitative RNA sequencing data from mouse hearts, livers, and skin cells, we identified 321 lncRNAs present in the heart, 117 of which exhibit a cardiac-enriched pattern of expression. By comparing lncRNA profiles of normal embryonic (∼E14), normal adult, and hypertrophied adult hearts, we defined a distinct fetal lncRNA abundance signature that includes 157 lncRNAs differentially expressed compared with adults (fold-change ≥ 50%, false discovery rate = 0. 02) and that was only poorly recapitulated in hypertrophied hearts (17 differentially expressed lncRNAs; 13 of these observed in embryonic hearts). Analysis of protein-coding mRNAs from the same samples identified 22 concordantly and 11 reciprocally regulated mRNAs within 10 kb of dynamically expressed lncRNAs, and reciprocal relationships of lncRNA and mRNA levels were validated for the Mccc1 and Relb genes using in vitro lncRNA knockdown in C2C12 cells. Network analysis suggested a central role for lncRNAs in modulating NFκB- and CREB1-regulated genes during embryonic heart growth and identified multiple mRNAs within these pathways that are also regulated, but independently of lncRNAs. VSports手机版.
Keywords: fetal heart; pressure overload. V体育安卓版.
VSports - Conflict of interest statement
The authors declare no conflict of interest.
Figures
References (VSports在线直播)
Publication types
MeSH terms
- "V体育官网入口" Actions
"V体育平台登录" Substances
- Actions (VSports在线直播)
Associated data
- "V体育官网" Actions
- V体育安卓版 - Actions
- Actions
- SRA/SRP026654
Grants and funding (V体育2025版)
LinkOut - more resources
Full Text Sources
Other Literature Sources
"VSports app下载" Molecular Biology Databases

