RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts
- PMID: 24105322
- PMCID: PMC4852478
- DOI: "V体育官网入口" 10.1038/nrm3679
RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts (VSports)
Abstract (V体育平台登录)
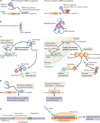
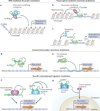
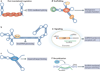
The increased application of transcriptome-wide profiling approaches has led to an explosion in the number of documented long non-coding RNAs (lncRNAs) VSports手机版. While these new and enigmatic players in the complex transcriptional milieu are encoded by a significant proportion of the genome, their functions are mostly unknown. Early discoveries support a paradigm in which lncRNAs regulate transcription via chromatin modulation, but new functions are steadily emerging. Given the biochemical versatility of RNA, lncRNAs may be used for various tasks, including post-transcriptional regulation, organization of protein complexes, cell-cell signalling and allosteric regulation of proteins. .
Figures

VSports最新版本 - References
-
-
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. Provides an overview of a series of papers releasee as part of the ENCODE project in which landmarks of biochemical function (regions of transcription, transcription factor association and histone modifications, among others) were attributed to 80% of the genome
-
-
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Koch E, Jourquin E, Ferrier P, Andrau J-C. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 2008;33:265–273. - PubMed
-
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. - PubMed
"V体育ios版" Publication types
MeSH terms
- "VSports手机版" Actions
- Actions (V体育官网入口)
- "VSports在线直播" Actions
Substances
- "V体育安卓版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

