V体育平台登录 - Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans
- PMID: 23846617
- PMCID: V体育2025版 - PMC3709164
- DOI: "V体育平台登录" 10.1038/srep02191
Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans (VSports注册入口)
"V体育平台登录" Abstract
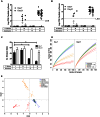
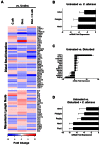
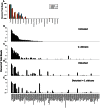
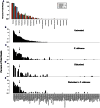
The introduction of Candida albicans into cefoperazone-treated mice results in changes in bacterial community reassembly. Our objective was to use high-throughput sequencing to characterize at much greater depth the specific changes in the bacterial microbiome. The colonization of C. albicans significantly altered bacterial community reassembly that was evident at multiple taxonomic levels of resolution. There were marked changes in the levels of Bacteriodetes and Lactobacillaceae. Lachnospiraceae and Ruminococcaceae, the two most abundant bacterial families, did not change in relative proportions after antibiotics, but there were marked genera-level shifts within these two bacterial families VSports手机版. The microbiome shifts occurred in the absence of overt intestinal inflammation. Overall, these experiments demonstrate that the introduction of a single new microbe in numerically inferior numbers into the bacterial microbiome during a broad community disturbance has the potential to significantly alter the subsequent reassembly of the bacterial community as it recovers from that disturbance. .
Figures


References
-
- Dethlefsen L., Huse S., Sogin M. L. & Relman D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6, e280 (2008). - PMC (VSports在线直播) - PubMed
-
- Jernberg C., Lofmark S., Edlund C. & Jansson J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal 1, 56–66 (2007). - PubMed
-
- Lofmark S., Jernberg C., Jansson J. K. & Edlund C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. The Journal of antimicrobial chemotherapy 58, 1160–1167 (2006). - PubMed
Publication types
- "VSports" Actions
MeSH terms
- Actions (VSports最新版本)
- "V体育ios版" Actions
- Actions (V体育2025版)
- "VSports注册入口" Actions
- Actions (VSports注册入口)
- "V体育2025版" Actions
- "V体育官网入口" Actions
- V体育安卓版 - Actions
- V体育官网 - Actions
- VSports - Actions
- VSports注册入口 - Actions
Substances
- "V体育官网" Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources

