V体育平台登录 - Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder
- PMID: 23375656
- PMCID: PMC3567267
- DOI: 10.1016/j.ajhg.2012.12.016
Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder
"V体育ios版" Abstract
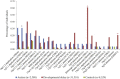
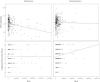
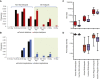
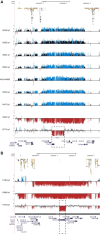
Rare copy-number variants (CNVs) have been implicated in autism and intellectual disability. These variants are large and affect many genes but lack clear specificity toward autism as opposed to developmental-delay phenotypes. We exploited the repeat architecture of the genome to target segmental duplication-mediated rearrangement hotspots (n = 120, median size 1. 78 Mbp, range 240 kbp to 13 Mbp) and smaller hotspots flanked by repetitive sequence (n = 1,247, median size 79 kbp, range 3-96 kbp) in 2,588 autistic individuals from simplex and multiplex families and in 580 controls. Our analysis identified several recurrent large hotspot events, including association with 1q21 duplications, which are more likely to be identified in individuals with autism than in those with developmental delay (p = 0. 01; OR = 2. 7). Within larger hotspots, we also identified smaller atypical CNVs that implicated CHD1L and ACACA for the 1q21 and 17q12 deletions, respectively. Our analysis, however, suggested no overall increase in the burden of smaller hotspots in autistic individuals as compared to controls VSports手机版. By focusing on gene-disruptive events, we identified recurrent CNVs, including DPP10, PLCB1, TRPM1, NRXN1, FHIT, and HYDIN, that are enriched in autism. We found that as the size of deletions increases, nonverbal IQ significantly decreases, but there is no impact on autism severity; and as the size of duplications increases, autism severity significantly increases but nonverbal IQ is not affected. The absence of an increased burden of smaller CNVs in individuals with autism and the failure of most large hotspots to refine to single genes is consistent with a model where imbalance of multiple genes contributes to a disease state. .
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc V体育安卓版. All rights reserved. .
V体育官网入口 - Figures


References (V体育官网入口)
-
- Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. - PubMed
-
- Sharp A.J., Cheng Z., Eichler E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. - PubMed (V体育ios版)
"V体育安卓版" Publication types
MeSH terms
- "V体育官网入口" Actions
- VSports app下载 - Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- V体育ios版 - Actions
- "V体育2025版" Actions
- "V体育官网" Actions
Associated data
- VSports - Actions
Grants and funding (V体育2025版)
LinkOut - more resources
Full Text Sources
"VSports注册入口" Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous (V体育2025版)

