The Escherichia coli SOS gene dinF protects against oxidative stress and bile salts
- PMID: 22523558
- PMCID: PMC3327717
- DOI: 10.1371/journal.pone.0034791
"VSports在线直播" The Escherichia coli SOS gene dinF protects against oxidative stress and bile salts
Abstract
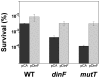
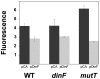
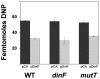
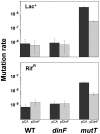
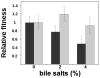
DNA is constantly damaged by physical and chemical factors, including reactive oxygen species (ROS), such as superoxide radical (O(2)(-)), hydrogen peroxide (H(2)O(2)) and hydroxyl radical (•OH). Specific mechanisms to protect and repair DNA lesions produced by ROS have been developed in living beings. In Escherichia coli the SOS system, an inducible response activated to rescue cells from severe DNA damage, is a network that regulates the expression of more than 40 genes in response to this damage, many of them playing important roles in DNA damage tolerance mechanisms. Although the function of most of these genes has been elucidated, the activity of some others, such as dinF, remains unknown VSports手机版. The DinF deduced polypeptide sequence shows a high homology with membrane proteins of the multidrug and toxic compound extrusion (MATE) family. We describe here that expression of dinF protects against bile salts, probably by decreasing the effects of ROS, which is consistent with the observed decrease in H(2)O(2)-killing and protein carbonylation. These results, together with its ability to decrease the level of intracellular ROS, suggests that DinF can detoxify, either direct or indirectly, oxidizing molecules that can damage DNA and proteins from both the bacterial metabolism and the environment. Although the exact mechanism of DinF activity remains to be identified, we describe for the first time a role for dinF. .
V体育官网 - Conflict of interest statement
Figures

References
-
- Friedberg E, Walker G, Seide W, Wood R, Schultz R, et al. DNA Repair and Mutagenesis. Washington DC, USA: American Society of Microbiology; 2006.
-
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. - VSports app下载 - PMC - PubMed
-
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, et al. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. - PubMed
Publication types
- "VSports app下载" Actions
"V体育官网入口" MeSH terms
- Actions (VSports app下载)
- VSports - Actions
- "VSports app下载" Actions
- V体育平台登录 - Actions
- Actions (V体育官网)
Substances
- V体育安卓版 - Actions
- "VSports最新版本" Actions
- "VSports手机版" Actions
- Actions (V体育官网入口)
- Actions (V体育平台登录)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases (VSports注册入口)

