V体育官网 - Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production
- PMID: 20702731
- PMCID: PMC3175530
- DOI: 10.4049/jimmunol.0903610
Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production
Abstract
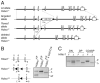
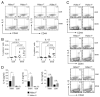
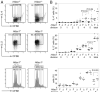
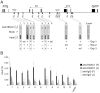
Chromatin modifications, such as reversible histone acetylation, play a key role in the regulation of T cell development and function. However, the role of individual histone deacetylases (HDACs) in T cells is less well understood. In this article, we show by conditional gene targeting that T cell-specific loss of HDAC1 led to an increased inflammatory response in an in vivo allergic airway inflammation model. Mice with HDAC1-deficient T cells displayed an increase in all critical parameters in this Th2-type asthma model, such as eosinophil recruitment into the lung, mucus hypersecretion, parenchymal lung inflammation, and enhanced airway resistance VSports手机版. This correlated with enhanced Th2 cytokine production in HDAC1-deficient T cells isolated from diseased mice. In vitro-polarized HDAC1-deficient Th2 cells showed a similar enhancement of IL-4 expression, which was evident already at day 3 of Th2 differentiation cultures and restricted to T cell subsets that underwent several rounds of cell divisions. HDAC1 was recruited to the Il4 gene locus in ex vivo isolated nonstimulated CD4(+) T cells, indicating a direct control of the Il4 gene locus. Our data provide genetic evidence that HDAC1 is an essential HDAC that controls the magnitude of an inflammatory response by modulating cytokine expression in effector T cells. .
Figures



References
-
- Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2002;2:909–919. - "V体育安卓版" PubMed
-
- Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv. Immunol. 2004;83:55–89. - V体育安卓版 - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. - "VSports手机版" PubMed
-
- Krangel MS. T cell development: better living through chromatin. Nat. Immunol. 2007;8:687–694. - PubMed
-
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. - "V体育2025版" PubMed
Publication types
"VSports app下载" MeSH terms
- V体育安卓版 - Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- Actions (VSports手机版)
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- Actions (VSports)
- VSports app下载 - Actions
- V体育官网入口 - Actions
- "V体育安卓版" Actions
Substances
- V体育2025版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

