The 3' end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein
- PMID: 14557646
- PMCID: PMC229252
- DOI: "V体育官网入口" 10.1128/jvi.77.21.11603-11615.2003
The 3' end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein
Abstract
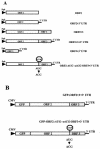
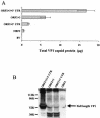
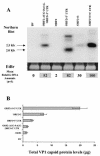
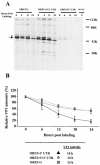
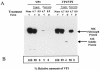
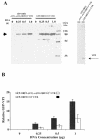
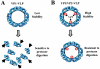
Norwalk virus (NV) is the prototype strain of a group of noncultivable human caliciviruses responsible for epidemic outbreaks of acute gastroenteritis. The capsid protein VP1 is synthesized from a subgenomic RNA that contains two open reading frames (ORFs), ORF2 and ORF3, and the 3' untranslated region (UTR). ORF2 and ORF3 code for the capsid protein (VP1) and a small structural basic protein (VP2), respectively. We discovered that the yields of virus-like particles (VLPs) composed of VP1 are significantly reduced when this protein is expressed from ORF2 alone VSports手机版. To determine how the 3' terminus of the NV subgenomic RNA regulates VP1 expression, we compared VP1 expression levels by using recombinant baculovirus constructs containing different 3' elements. High VP1 levels were detected by using a recombinant baculovirus that contained ORF2, ORF3, and the 3'UTR (ORF2+3+3'UTR). In contrast, expression of VP1 from constructs that lacked the 3'UTR (ORF2+3), ORF3 (ORF2+3'UTR), or both (ORF2 alone) was highly reduced. Elimination of VP2 synthesis from the subgenomic RNA by mutation resulted in VP1 levels similar to those obtained with the ORF2 construct alone, suggesting a cis role for VP2 in upregulation of VP1 expression levels. Comparisons of the kinetics of RNA and capsid protein expression levels by using constructs with or without ORF3 or the 3'UTR revealed that the 3'UTR increased the levels of VP1 RNA, whereas the presence of VP2 resulted in increased levels of VP1. Furthermore, VP2 increased VP1 stability and protected VP1 from disassembly and protease degradation. The increase in VP1 expression levels caused by the presence of VP2 in cis was also observed in mammalian cells. .
Figures
"V体育安卓版" References
-
- Agrawal, S., D. Gupta, and S. K. Panda. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87-101. - PubMed
-
- Arora, R., C. Priano, A. B. Jacobson, and D. R. Mills. 1996. cis-Acting elements within an RNA coliphage genome: fold as you please, but fold you must!! J. Mol. Biol. 258:433-446. - PubMed
-
- Belliot, G., J. S. Noel, J.-F. Li, S. Yoshiyuki, Humphrey, T. M., Ando, T., Glass, R. I., and Monroe, S. S. 2001. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses.” J. Clin. Microbiol. 39:4288-4294. - V体育2025版 - PMC - PubMed
Publication types
- "V体育官网入口" Actions
MeSH terms (VSports在线直播)
- Actions (V体育安卓版)
- Actions (V体育平台登录)
- V体育2025版 - Actions
- "VSports app下载" Actions
- "V体育官网入口" Actions
- V体育官网入口 - Actions
Substances
- "V体育ios版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources (VSports最新版本)

