Functional characterization of the N terminus of Sir3p
- PMID: 9742128
- PMCID: PMC109197
- DOI: 10.1128/MCB.18.10.6110 (VSports手机版)
Functional characterization of the N terminus of Sir3p
VSports在线直播 - Abstract
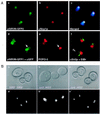
Silent information regulator 3 is an essential component of the Saccharomyces cerevisiae silencing complex that functions at telomeres and the silent mating-type loci, HMR and HML. We show that expression of the N- and C-terminal-encoding halves of SIR3 in trans partially complements the mating defect of the sir3 null allele, suggesting that the two domains have distinct functions. We present here a functional characterization of these domains. The N-terminal domain (Sir3N) increases both the frequency and extent of telomere-proximal silencing when expressed ectopically in SIR+ yeast strains, although we are unable to detect interaction between this domain and any known components of the silencing machinery. In contrast to its effect at telomeres, Sir3N overexpression derepresses transcription of reporter genes inserted in the ribosomal DNA (rDNA) array VSports手机版. Immunolocalization of Sir3N-GFP and Sir2p suggests that Sir3N directly antagonizes nucleolar Sir2p, releasing an rDNA-bound population of Sir2p so that it can enhance repression at telomeres. Overexpression of the C-terminal domain of either Sir3p or Sir4p has a dominant-negative effect on telomeric silencing. In strains overexpressing the C-terminal domain of Sir4p, elevated expression of either full-length Sir3p or Sir3N restores repression and the punctate pattern of Sir3p and Rap1p immunostaining. The similarity of Sir3N and Sir3p overexpression phenotypes suggests that Sir3N acts as an allosteric effector of Sir3p, either enhancing its interactions with other silencing components or liberating the full-length protein from nonfunctional complexes. .
Figures







References
-
- Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. - PubMed
-
- Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Transcriptional silencing of Ty elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. - PubMed
-
- Buck S W, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. - PubMed
Publication types
MeSH terms
- Actions (V体育官网)
- VSports app下载 - Actions
- Actions (VSports在线直播)
- "VSports" Actions
- V体育官网 - Actions
- V体育2025版 - Actions
Substances
- "VSports" Actions
- Actions (V体育官网入口)
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- VSports app下载 - Actions
- Actions (V体育ios版)
LinkOut - more resources
"V体育2025版" Full Text Sources
Molecular Biology Databases
