Prognostic and Immunological Significance of the Molecular Subtypes and Risk Signatures Based on Cuproptosis in Hepatocellular Carcinoma
- PMID: 37124062
- PMCID: PMC10139815 (VSports手机版)
- DOI: 10.1155/2023/3951940 (VSports app下载)
Prognostic and Immunological Significance of the Molecular Subtypes and Risk Signatures Based on Cuproptosis in Hepatocellular Carcinoma (V体育官网入口)
Abstract
Background: Hepatocellular carcinoma (HCC) remains a challenging medical problem. Cuproptosis is a novel form of cell death that plays a crucial role in tumorigenesis, angiogenesis, and metastasis. However, it remains unclear whether cuproptosis-related genes (CRGs) influence the outcomes and immune microenvironment of HCC patients VSports手机版. .
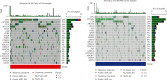
Method: From The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases, we obtained the mRNA expression file and related clinical information of HCC patients. We selected 19 CRGs as candidate genes for this study according to previous literature. We performed a differential expression analysis of the 19 CRGs between malignant and precancerous tissue V体育安卓版. Based on the 19 CRGs, we enrolled cluster analysis to identify cuproptosis-related subtypes of HCC patients. A prognostic risk signature was created utilizing univariate Cox regression and least absolute shrinkage and selection operator (LASSO) regression analyses. We employed independent and stratification survival analyses to investigate the predictive value of this model. The functional enrichment features, mutation signatures, immune profile, and response to immunotherapy of HCC patients were also investigated according to the two molecular subtypes and the prognostic signature. .
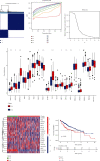
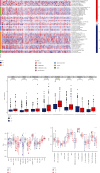
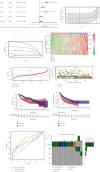
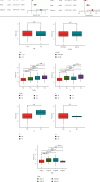
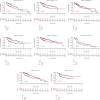
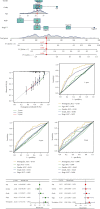
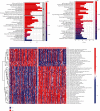
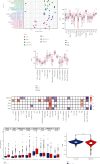
Results: We found that 17 CRGs significantly differed in HCC versus normal samples. Cluster analysis showed two distinct molecular subtypes of cuproptosis. Cluster 1 is preferentially related to poor prognosis, high activity of immune response signaling, high mutant frequency of TP53, and distinct immune cell infiltration versus cluster 2 V体育ios版. Through univariate and LASSO Cox regression analyses, we created a cuproptosis-related prognostic risk signature containing LIPT1, DLAT, MTF1, GLS, and CDKN2A. High-risk HCC patients were shown to have a worse prognosis. The risk signature was proved to be an independent predictor of prognosis in both the TCGA and ICGC datasets, according to multivariate analysis. The signature also performed well in different stratification of clinical features. The immune cells, which included regulatory T cells (Treg), B cells, macrophages, mast cells, NK cells, and aDCs, as well as immune functions containing cytolytic activity, MHC class I, and type II IFN response, were remarkably distinct between the high-risk and low-risk groups. The tumor immune dysfunction and exclusion (TIDE) score suggested that high-risk patients had a higher response rate to immune checkpoint inhibitors than low-risk patients. .
Conclusion: This research discovered the potential prognostic and immunological significance of cuproptosis in HCC, improved the understanding of cuproptosis, and may deliver new directions for developing more efficacious therapeutic techniques for HCC patients VSports最新版本. .
Copyright © 2023 Xiaolong Tang et al.
Conflict of interest statement
No potential conflicts of interest were disclosed.
VSports最新版本 - Figures



References
-
- Llovet J. M., Ricci S., Mazzaferro V., et al. Sorafenib in advanced hepatocellular carcinoma. The New England Journal of Medicine . 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. - DOI (V体育ios版) - PubMed
MeSH terms
- V体育官网入口 - Actions
- "VSports注册入口" Actions
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions (VSports最新版本)
- V体育官网 - Actions
- V体育官网 - Actions
- "V体育安卓版" Actions
- V体育2025版 - Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
Substances
LinkOut - more resources
VSports手机版 - Full Text Sources
Medical
Research Materials
Miscellaneous

