Revealing Prognostic and Immunotherapy-Sensitive Characteristics of a Novel Cuproptosis-Related LncRNA Model in Hepatocellular Carcinoma Patients by Genomic Analysis (VSports app下载)
- PMID: 36672493
- PMCID: PMC9857215
- DOI: 10.3390/cancers15020544
VSports最新版本 - Revealing Prognostic and Immunotherapy-Sensitive Characteristics of a Novel Cuproptosis-Related LncRNA Model in Hepatocellular Carcinoma Patients by Genomic Analysis
Abstract
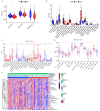
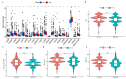
Immunotherapy has shown strong anti-tumor activity in a subset of patients. However, many patients do not benefit from the treatment, and there is no effective method to identify sensitive immunotherapy patients. Cuproptosis as a non-apoptotic programmed cell death caused by excess copper, whether it is related to tumor immunity has attracted our attention. In the study, we constructed the prognostic model of 9 cuproptosis-related LncRNAs (crLncRNAs) and assessed its predictive capability, preliminarily explored the potential mechanism causing treatment sensitivity difference between the high-/low-risk group. Our results revealed that the risk score was more effective than traditional clinical features in predicting the survival of HCC patients (AUC = 0. 828). The low-risk group had more infiltration of immune cells (B cells, CD8+ T cells, CD4+ T cells), mainly with anti-tumor immune function (p < 0. 05). It showed higher sensitivity to immune checkpoint inhibitors (ICIs) treatment (p < 0. 001) which may exert the effect through the AL365361. 1/hsa-miR-17-5p/NLRP3 axis. In addition, NLRP3 mutation-sensitive drugs (VNLG/124, sunitinib, linifanib) may have better clinical benefits in the high-risk group. All in all, the crLncRNAs model has excellent specificity and sensitivity, which can be used for classifying the therapy-sensitive population and predicting the prognosis of HCC patients. VSports手机版.
Keywords: AL365361. 1; cuproptosis-related LncRNAs (crLncRNAs); immune checkpoint inhibitors (ICIs); immunotherapy; tumor immune microenvironment (TIME). V体育安卓版.
Conflict of interest statement
The authors declare no conflict of interest.
"VSports" Figures





References
-
- Finn R.S., Ryoo B.-Y., Merle P., Kudo M., Bouattour M., Lim H.Y., Breder V., Edeline J., Chao Y., Ogasawara S., et al. Pembrolizumab as Second-Line Therapy in Patients with Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020;38:193–202. doi: 10.1200/JCO.19.01307. - V体育2025版 - DOI - PubMed
"VSports注册入口" Grants and funding
- V体育ios版 - No. 81903149/National Natural Science Foundation of China
- No. 2021SF-058/Natural Science Basic Research Project of Shaanxi Province, China
- No. 2019YW19/Shaanxi Province "High-level Talents Special Support Program" project (Wenjie Song) and the Medical and Health science and technology Foundation of Sanya
LinkOut - more resources
"V体育官网入口" Full Text Sources
Research Materials

