Multi-omics pan-cancer study of cuproptosis core gene FDX1 and its role in kidney renal clear cell carcinoma
- PMID: 36605188
- PMCID: PMC9810262
- DOI: 10.3389/fimmu.2022.981764
Multi-omics pan-cancer study of cuproptosis core gene VSports注册入口 - FDX1 and its role in kidney renal clear cell carcinoma
Abstract
Background: The mechanism of copper-induced cellular death was newly discovered and termed cuproptosis VSports手机版. Inducing cuproptosis in cancer cells is well anticipated for its curative potential in treating tumor diseases. However, ferredoxin 1 (FDX1), the core regulatory gene in cuproptosis, is rarely studied, and the regulation of FDX1 in tumor biology remains obscure. A comprehensive pan-cancer analysis of FDX1 is needed. .
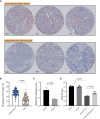
Methods: Thirty-three types of tumors were included with paired normal tissues in The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) datasets. The interaction between transcription, protein, phosphorylation, and promoter methylation levels was analyzed. Survival, immune infiltration, single-cell FDX1 expression, FDX1-related tumor mutational burden (TMB), microsatellite instability (MSI), stemness, tumor immune dysfunction and exclusion (TIDE), and immunotherapy-related analyses were performed V体育安卓版. FDX1 protein expression was assessed by kidney renal clear cell carcinoma (KIRC) tissue microarray immunohistochemistry. The function of FDX1 in KIRC was further explored by experiments in 786-O cell lines in vitro. .
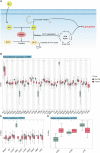
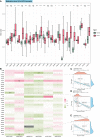
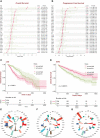
Results: FDX1 is highly expressed in 15 tumor types and lowly expressed in 11 tumor types. The corresponding changes in protein expression, phosphorylation, and promoter methylation level of FDX1 have been described in several tumors. Survival analysis showed that FDX1 was related to favorable or poor overall survival in eight tumors and progression-free survival in nine tumors. Immune infiltration and single-cell analysis indicated the indispensable role of FDX1 expression in macrophages and monocytes. Multiple established immunotherapy cohorts suggested that FDX1 may be a potential predictor of treatment effects for tumor patients. Tissue microarray analysis showed decreased FDX1 expression in KIRC patients' tumor tissues. Knockdown of FDX1 resulted in the downregulation of cuproptosis in kidney renal clear tumor cells. Mechanistically, the FDX1-associated gene expression signature in KIRC is related to the enrichment of genes involved in the tricarboxylic acid (TCA) cycle, NOTCH pathway, etc. Several NOTCH pathway genes were differentially expressed in the high- and low-FDX1 groups in KIRC V体育ios版. .
Conclusion: Our analysis showed that the central regulatory gene of cuproptosis, FDX1, has differential expression and modification levels in various tumors, which is associated with cellular function, immune modulation, and disease prognosis VSports最新版本. Thus, FDX1-dependent cuproptosis may serve as a brand-new target in future therapeutic approaches against tumors. .
Keywords: FDX1; KIRC; cuproptosis; immunology; notch; pan-cancer V体育平台登录. .
Copyright © 2022 Xu, Hu, Cao, Zhang, Luo, Zhang, Wang, Cheng and Li VSports注册入口. .
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest V体育官网入口. The reviewer LS declared a shared affiliation with the authors JX, ZH, HZ, XW, QC at the time of review.
Figures




"V体育官网入口" References
-
- Sheftel AD, Stehling O, Pierik AJ, Elsässer HP, Mühlenhoff U, Webert H, et al. . Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA (2010) 107(26):11775–80. doi: 10.1073/pnas.1004250107 - DOI - PMC - PubMed
-
- Wang Z, Dong H, Yang L, Yi P, Wang Q, Huang D. The role of Fdx1 in granulosa cell of polycystic ovary syndrome (Pcos). BMC Endocr Disord (2021) 21(1):119. doi: 10.1186/s12902-021-00775-w - VSports - DOI - PMC - PubMed
"VSports最新版本" Publication types
- "V体育ios版" Actions
MeSH terms
- "V体育2025版" Actions
- VSports注册入口 - Actions
- V体育官网入口 - Actions
- V体育平台登录 - Actions
- VSports最新版本 - Actions
Substances
- Actions (VSports app下载)
- "VSports手机版" Actions
LinkOut - more resources
Full Text Sources
Medical

