Molecular subtypes based on cuproptosis regulators and immune infiltration in kidney renal clear cell carcinoma (V体育2025版)
- PMID: 36338990
- PMCID: PMC9635053
- DOI: 10.3389/fgene.2022.983445 (V体育ios版)
V体育官网入口 - Molecular subtypes based on cuproptosis regulators and immune infiltration in kidney renal clear cell carcinoma
Abstract
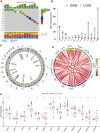
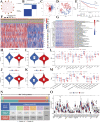
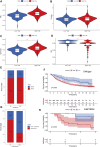
Copper toxicity involves the destruction of mitochondrial metabolic enzymes, triggering an unusual mechanism of cell death called cuproptosis, which proposes a novel approach using copper toxicity to treat cancer. However, the biological function of cuproptosis has not been fully elucidated in kidney renal clear cell carcinoma (KIRC). Using the expression profile of 13 cuproptosis regulators, we first identified two molecular subtypes related to cuproptosis defined as "hot tumor" and "cold tumor", having different levels of biological function, clinical prognosis, and immune cell infiltration. We obtained three gene clusters using the differentially expressed genes between the two cuproptosis-related subtypes, which were associated with different molecular activities and clinical characteristics. Next, we developed and validated a cuproptosis prognostic model that included two genes (FDX1 and DBT). The calculated risk score could divide patients into high- and low-risk groups. The high-risk group had a poorer prognosis, lower level of immune infiltration, higher frequency of gene alterations, and greater levels of FDX1 methylation and limited DBT methylation. The risk score was also an independent predictive factor for overall survival in KIRC. The established nomogram calculating the risk score achieved a high predictive ability for the prognosis of individual patients (area under the curve: 0. 860). We then identified small molecular inhibitors as potential treatments and analyzed the sensitivity to chemotherapy of the signature genes VSports手机版. Tumor immune dysfunction and exclusion (TIDE) showed that the high-risk group had a higher level of TIDE, exclusion and dysfunction that was lower than the low-risk group, while the microsatellite instability of the high-risk group was significantly lower. The results of two independent immunotherapy datasets indicated that cuproptosis regulators could influence the response and efficacy of immunotherapy in KIRC. Our study provides new insights for individualized and comprehensive therapy of KIRC. .
Keywords: KIRC; cuproptosis; immune infiltration; immunotherapy; molecular subtypes. V体育安卓版.
Copyright © 2022 Liu, Li, Shen, Li, Zhao, Shen and Li V体育ios版. .
Conflict of interest statement (VSports)
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Capitanio U., Bensalah K., Bex A., Boorjian S. A., Bray F., Coleman J., et al. (2019). Epidemiology of renal cell carcinoma. Eur. Urol. 75, 74–84. 10.1016/j.eururo.2018.08.036 - V体育ios版 - DOI - PMC - PubMed
V体育官网入口 - LinkOut - more resources
V体育官网 - Full Text Sources

