"VSports" Paired Donor and Recipient Immunophenotyping in Allogeneic Hematopoietic Stem Cell Transplantation: A Cellular Network Approach
- PMID: 35677053
- PMCID: PMC9168993
- DOI: 10.3389/fimmu.2022.874499
Paired Donor and Recipient Immunophenotyping in Allogeneic Hematopoietic Stem Cell Transplantation: A Cellular Network Approach
"V体育ios版" Abstract
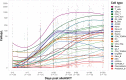
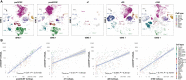
Success and complications of allogeneic hematopoietic stem cell transplantation (alloHSCT) are closely connected to the transferred graft and immune reconstitution post alloHSCT. Due to the variety of immune cells and their distinct roles, a broad evaluation of the immune cellular network is warranted in mobilization and reconstitution studies in alloHSCT. Here, we propose a comprehensive phenotypic analysis of 26 immune cell subsets with multicolor flow cytometry from only 100µl whole blood per time point VSports手机版. Using this approach, we provide an extensive longitudinal analysis of almost 200 time points from 21 donor-recipient pairs. We observe a broad mobilization of innate and adaptive immune cell subsets after granulocyte-colony stimulating factor (G-CSF) treatment of healthy donors. Our data suggest that the relative quantitative immune cell subset composition in recipients approaches that of healthy donors from day +180 post alloHSCT onwards. Correlation of donor and recipient cell counts reveals distinct association patterns for different immune cell subsets and hierarchical clustering of recipient cell counts identifies distinct reconstitution groups in the first month after transplantation. We suggest our comprehensive immune subset analysis as a feasible and time efficient approach for a broad immune assessment for future clinical studies in the context of alloHSCT. This comprehensive cell composition assessment can be a critical step towards personalized graft composition strategies and individualized therapy management in areas such as GvHD prophylaxis in the highly complex immunological setting of alloHSCT. .
Keywords: allogeneic hematopoietic stem cell transplantation (HSCT); granulocyte-colony stimulating factor (G-CSF); immune cell network; immune subsets; mobilization; reconstitution V体育安卓版. .
Copyright © 2022 Wittenbecher, Lesch, Kolling, Blau, Vuong, Borchert, Movasshagi, Tietze-Bürger, Penack, Ahn, Bullinger, Frentsch and Na V体育ios版. .
Conflict of interest statement
The authors declare that the research for this study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest VSports最新版本. The reviewer HTG declared past co-authorships with the author OP to the handling editor.
Figures



"VSports在线直播" References
-
- Singh AK, McGuirk JP. Allogeneic Stem Cell Transplantation: A Historical and Scientific Overview. Cancer Res (2016) 76(22):6445–51. doi: 10.1158/0008-5472.CAN-16-1311 - "VSports注册入口" DOI - PubMed
-
- Storek J, Dawson MA, Storer B, Stevens-Ayers T, Maloney DG, Marr KA, et al. . Immune Reconstitution After Allogeneic Marrow Transplantation Compared With Blood Stem Cell Transplantation. Blood (2001) 97(11):3380–9. doi: 10.1182/blood.V97.11.3380 - DOI (V体育平台登录) - PubMed
-
- Melve GK, Ersvaer E, Eide GE, Kristoffersen EK, Bruserud Ø. Peripheral Blood Stem Cell Mobilization in Healthy Donors by Granulocyte Colony-Stimulating Factor Causes Preferential Mobilization of Lymphocyte Subsets. Front Immunol (2018) 9:845. doi: 10.3389/fimmu.2018.00845 - V体育2025版 - DOI - PMC - PubMed
Publication types
- Actions (V体育2025版)
MeSH terms
- Actions (VSports app下载)
- Actions (V体育平台登录)
- Actions (VSports在线直播)
Substances (VSports)
LinkOut - more resources
Full Text Sources

