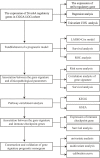
Establishment and Validation of a 5 m6A RNA Methylation Regulatory Gene Prognostic Model in Low-Grade Glioma (V体育2025版)
- PMID: 35281815
- PMCID: PMC8914514
- DOI: 10.3389/fgene.2022.655169
Establishment and Validation of a 5 m6A RNA Methylation Regulatory Gene Prognostic Model in Low-Grade Glioma
Abstract
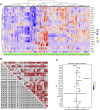
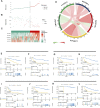
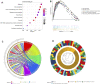
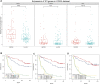
Background: The prognosis of low-grade glioma (LGG) is different from that of other intracranial tumors. Although many markers of LGG have been established, few are used in clinical practice. M6A methylation significantly affects the biological behavior of LGG tumors. Therefore, establishment of an LGG prognostic model based on m6A methylation regulatory genes is of great interest. Methods: Data from 495 patients from The Cancer Genome Atlas (TCGA) and 172 patients from the Chinese Glioma Genome Atlas (CGGA) were analyzed. Univariate Cox analysis was used to identify methylation regulatory genes with prognostic significance. LASSO Cox regression was used to identify prognostic genes. Receiver operating characteristic (ROC) and Kaplan-Meier curves were used to verify the accuracy of the model. Gene Set Enrichment Analysis (GSEA) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to identify cellular pathways that were significantly associated with the prognosis of LGG. Results: A glioma prognostic model based on five methylation regulatory genes was established. Compared with low-risk patients, patients identified as high risk had a poorer prognosis VSports手机版. There was a high degree of consistency between the internal training and internal validation CGGA cohorts and the external validation TCGA cohort. Furthermore, KEGG and GSEA analyses showed that the focal adhesion and cell cycle pathways were significantly upregulated in high-risk patients. This signature could be used to distinguish among patients with different immune checkpoint gene expression levels, which may inform immune checkpoint inhibitor (ICI) immunotherapy. Conclusion: We comprehensively evaluated m6A methylation regulatory genes in LGG and constructed a prognostic model based on m6A methylation, which may improve prognostic prediction for patients with LGG. .
Keywords: ICI; LASSO; LGG; M6A RNA methylation; prognostic model. V体育安卓版.
Copyright © 2022 Bai, Wang and Zhang.
"V体育官网入口" Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alarcón C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F. (2015). N6-methyladenosine marks Primary microRNAs for Processing. Nature 519 (7544), 482–485. 10.1038/nature14281 - VSports在线直播 - DOI - PMC - PubMed
-
- Brat D. J., Brat D. J., Verhaak R. G., Aldape K. D., Yung W. K., Salama S. R., et al. (2015). Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 372 (26), 2481–2498. 10.1056/NEJMoa1402121 - "VSports在线直播" DOI - PMC - PubMed
-
- Chen P.-Y., Li X.-D., Ma W.-N., Li H., Li M.-M., Yang X.-Y., et al. (2020). Comprehensive Transcriptomic Analysis and Experimental Validation Identify lncRNA HOXA-AS2/miR-184/COL6A2 as the Critical ceRNA Regulation Involved in Low-Grade Glioma Recurrence. Ott 13, 4999–5016. 10.2147/ott.S245896 - VSports - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

