VSports注册入口 - The Landscape of Featured Metabolism-Related Genes and Imbalanced Immune Cell Subsets in Sepsis
- PMID: 35265105
- PMCID: PMC8901109
- DOI: 10.3389/fgene.2022.821275
The Landscape of Featured Metabolism-Related Genes and Imbalanced Immune Cell Subsets in Sepsis
"V体育ios版" Abstract
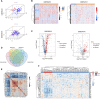
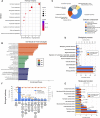
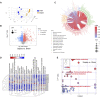
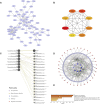
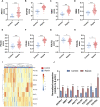
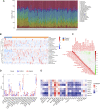
Sepsis is a heterogeneous disease state triggered by an uncontrolled inflammatory host response with high mortality and morbidity in severely ill patients. Unfortunately, the treatment effectiveness varies among sepsis patients and the underlying mechanisms have yet to be elucidated. The present aim is to explore featured metabolism-related genes that may become the biomarkers in patients with sepsis. In this study, differentially expressed genes (DEGs) between sepsis and non-sepsis in whole blood samples were identified using two previously published datasets (GSE95233 and GSE54514) VSports手机版. A total of 66 common DEGs were determined, namely, 52 upregulated and 14 downregulated DEGs. The Gene Set Enrichment Analysis (GSEA) results indicated that these DEGs participated in several metabolic processes including carbohydrate derivative, lipid, organic acid synthesis oxidation reduction, and small-molecule biosynthesis in patients with sepsis. Subsequently, a total of 8 hub genes were screened in the module with the highest score from the Cytoscape plugin cytoHubba. Further study showed that these hub DEGs may be robust markers for sepsis with high area under receiver operating characteristic curve (AUROC). The diagnostic values of these hub genes were further validated in myocardial tissues of septic rats and normal controls by untargeted metabolomics analysis using liquid chromatography-mass spectrometry (LC-MS). Immune cell infiltration analysis revealed that different infiltration patterns were mainly characterized by B cells, T cells, NK cells, monocytes, macrophages, dendritics, eosinophils, and neutrophils between sepsis patients and normal controls. This study indicates that metabolic hub genes may be hopeful biomarkers for prognosis prediction and precise treatment in sepsis patients. .
Keywords: bioinformatics; biomarkers; immune cell infiltration; metabolomics; sepsis. V体育安卓版.
Copyright © 2022 She, Tan, Zhou, Zhu, Ma, Wu, Du, Liu, Hu, Mao and Li. V体育ios版.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
"VSports注册入口" Figures

References
-
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. 10.1016/j.molp.2020.06.009 - VSports app下载 - DOI - PubMed
-
- Cho J. H., Ju W. S., Seo S. Y., Kim B. H., Kim J.-S., Kim J.-G., et al. (2021). The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor. Ijms 22 (22), 12194. 10.3390/ijms222212194 - VSports最新版本 - DOI - PMC - PubMed
LinkOut - more resources (V体育ios版)
Full Text Sources

