Vulnerability of Triple-Negative Breast Cancer to Saponin Formosanin C-Induced Ferroptosis
- PMID: 35204181
- PMCID: PMC8868405
- DOI: 10.3390/antiox11020298
VSports在线直播 - Vulnerability of Triple-Negative Breast Cancer to Saponin Formosanin C-Induced Ferroptosis
"V体育ios版" Abstract
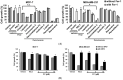
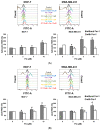
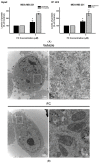
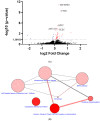
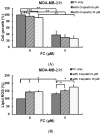
Targeting ferritin via autophagy (ferritinophagy) to induce ferroptosis, an iron- and reactive oxygen species (ROS)-dependent cell death, provides novel strategies for cancer therapy VSports手机版. Using a ferroptosis-specific inhibitor and iron chelator, the vulnerability of triple-negative breast cancer (TNBC) MDA-MB-231 cells to ferroptosis was identified and compared to that of luminal A MCF-7 cells. Saponin formosanin C (FC) was revealed as a potent ferroptosis inducer characterized by superior induction in cytosolic and lipid ROS formation as well as GPX4 depletion in MDA-MB-231 cells. The FC-induced ferroptosis was paralleled by downregulation of ferroportin and xCT expressions. Immunoprecipitation and electron microscopy demonstrated the involvement of ferritinophagy in FC-treated MDA-MB-231 cells. The association of FC with ferroptosis was strengthened by the results that observed an enriched pathway with differentially expressed genes from FC-treated cells. FC sensitized cisplatin-induced ferroptosis in MDA-MB-231 cells. Through integrated analysis of differentially expressed genes and pathways using the METABRIC patients' database, we confirmed that autophagy and ferroptosis were discrepant between TNBC and luminal A and that TNBC was hypersensitive to ferroptosis. Our data suggest a therapeutic strategy by ferroptosis against TNBC, an aggressive subtype with a poor prognosis. .
Keywords: breast cancer; ferritinophagy; ferroptosis potential index; formosanin C; gene database. V体育安卓版.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







VSports手机版 - References
-
- Nunez Abad M., Calabuig-Farinas S., Lobo de Mena M., Jose Godes Sanz de Bremond M., Garcia Gonzalez C., Torres Martinez S., Garcia-Garcia J.A., Iranzo Gonzalez-Cruz V., Camps Herrero C. Update on systemic treatment in early triple negative breast cancer. Ther. Adv. Med. Oncol. 2021;13:1758835920986749. doi: 10.1177/1758835920986749. - DOI - PMC - PubMed
-
- Dwivedi S., Purohit P., Misra R., Lingeswaran M., Vishnoi J.R., Pareek P., Misra S., Sharma P. Single Cell Omics of Breast Cancer: An Update on Characterization and Diagnosis. Indian J. Clin. Biochem. 2019;34:3–18. doi: 10.1007/s12291-019-0811-0. - DOI (V体育官网) - PMC - PubMed
VSports app下载 - Grants and funding
V体育官网 - LinkOut - more resources
"V体育安卓版" Full Text Sources
Miscellaneous (V体育平台登录)

