"VSports在线直播" Identification of Hub Genes and Potential ceRNA Networks of Diabetic Nephropathy by Weighted Gene Co-Expression Network Analysis
- PMID: 34790229
- PMCID: PMC8591079
- DOI: "VSports" 10.3389/fgene.2021.767654
Identification of Hub Genes and Potential ceRNA Networks of Diabetic Nephropathy by Weighted Gene Co-Expression Network Analysis
"VSports在线直播" Abstract
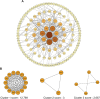
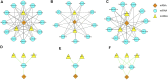
Diabetic nephropathy (DN) is one of the most common microvascular complications in diabetic patients, and is the main cause of end-stage renal disease. The exact molecular mechanism of DN is not fully understood. The aim of this study was to identify novel biomarkers and mechanisms for DN disease progression by weighted gene co-expression network analysis (WGCNA). From the GSE142153 dataset based on the peripheral blood monouclear cells (PBMC) of DN, we identified 234 genes through WGCNA and differential expression analysis VSports手机版. Gene Ontology (GO) annotations mainly included inflammatory response, leukocyte cell-cell adhesion, and positive regulation of proteolysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways mostly included IL-17 signaling pathway, MAPK signaling pathway, and PPAR signaling pathway in DN. A total of four hub genes (IL6, CXCL8, MMP9 and ATF3) were identified by cytoscape, and the relative expression levels of hub genes were also confirmed by RT-qPCR. ROC curve analysis determined that the expression of the four genes could distinguish DN from controls (the area under the curve is all greater than 0. 8), and Pearson correlation coefficient analysis suggested that the expression of the four genes was related to estimated glomerular filtration rate (eGFR) of DN. Finally, through database prediction and literature screening, we constructed lncRNA-miRNA-mRNA network. We propose that NEAT1/XIST/KCNQ1T1-let-7b-5p-IL6, NEAT1/XIST-miR-93-5p-CXCL8 and NEAT1/XIST/KCNQ1T1-miR-27a-3p/miR-16-5p-ATF3 might be potential RNA regulatory pathways to regulate the disease progression of early DN. In conclusion, we identified four hub genes, namely, IL6, CXCL8, MMP9, and ATF3, as markers for early diagnosis of DN, and provided insight into the mechanisms of disease development in DN at the transcriptome level. .
Keywords: RNA regulatory pathways; RT-qPCR; diabetic nephropathy; hub gene; weighted gene co-expression network analysis V体育安卓版. .
Copyright © 2021 Li, Zhang, Liu, Wei, Wang, Lv, Ye, Liu and Li. V体育ios版.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Assmann T. S., Recamonde-Mendoza M., Costa A. R., Puñales M., Tschiedel B., Canani L. H., et al. (2019). Circulating miRNAs in Diabetic Kidney Disease: Case-Control Study and In Silico Analyses. Acta Diabetol. 56 (1), 55–65. 10.1007/s00592-018-1216-x - DOI (V体育官网入口) - PubMed
-
- Chin C.-H., Chen S.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. (2014). cytoHubba: Identifying Hub Objects and Sub-networks from Complex Interactome. BMC Syst. Biol. 8 (4), S11. 10.1186/1752-0509-8-s4-s11 - VSports app下载 - DOI - PMC - PubMed
"V体育安卓版" LinkOut - more resources
Full Text Sources
Research Materials
"VSports手机版" Miscellaneous

