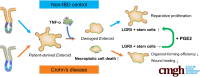
TNFα Induces LGR5+ Stem Cell Dysfunction In Patients With Crohn's Disease (V体育2025版)
- PMID: 34700029
- PMCID: PMC8783132
- DOI: 10.1016/j.jcmgh.2021.10.010
TNFα Induces LGR5+ Stem Cell Dysfunction In Patients With Crohn's Disease (V体育安卓版)
Abstract
Background & aims: Tumor necrosis factor alpha (TNFα) is considered a major tissue damage-promoting effector in Crohn's disease (CD) pathogenesis. Patient-derived intestinal organoid (enteroid) recapitulates the disease-specific characteristics of the intestinal epithelium VSports手机版. This study aimed to evaluate the intestinal epithelial responses to TNFα in enteroids derived from healthy controls and compare them with those of CD patient-derived enteroids. .
Methods: Human enteroids derived from patients with CD and controls were treated with TNFα (30 ng/mL), and cell viability and gene expression patterns were evaluated. V体育安卓版.
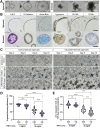
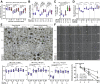
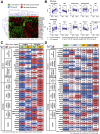
Results: TNFα induced MLKL-mediated necroptotic cell death, which was more pronounced in CD patient-derived enteroids than in control enteroids. Immunohistochemistry and RNA sequencing revealed that treatment with TNFα caused expansion of the intestinal stem cell (ISC) populations V体育ios版. However, expanded ISC subpopulations differed in control and CD patient-derived enteroids, with LGR5+ active ISCs in control enteroids and reserve ISCs, such as BMI1+ cells, in CD patient-derived enteroids. In single-cell RNA sequencing, LGR5+ ISC-enriched cell cluster showed strong expression of TNFRSF1B (TNFR2) and cyclooxygenase-prostaglandin E2 (PGE2) activation. In TNFα-treated CD patient-derived enteroids, exogenous PGE2 (10 nmol/L) induced the expansion of the LGR5+ ISC population and improved organoid-forming efficiency, viability, and wound healing. .
Conclusions: TNFα increases necroptosis of differentiated cells and induces the expansion of LGR5+ ISCs. In CD patient-derived enteroids, TNFα causes LGR5+ stem cell dysfunction (expansion failure), and exogenous PGE2 treatment restored the functions of LGR5+ stem cells VSports最新版本. Therefore, PGE2 can be used to promote mucosal healing in patients with CD. .
Keywords: Crohn’s Disease; Intestinal Organoid; Intestinal Stem Cell; Prostaglandin E2; Tumor Necrosis Factor-Alpha V体育平台登录. .
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved VSports注册入口. .
"VSports app下载" Figures






References
-
- Pizarro T.T., Stappenbeck T.S., Rieder F., Rosen M.J., Colombel J.F., Donowitz M., Towne J., Mazmanian S.K., Faith J.J., Hodin R.A., Garrett W.S., Fichera A., Poritz L.S., Cortes C.J., Shtraizent N., Honig G., Snapper S.B., Hurtado-Lorenzo A., Salzman N.H., Chang E.B. Challenges in IBD research: preclinical human IBD mechanisms. Inflamm Bowel Dis. 2019;25(Suppl 2):S5–S12. - PubMed
-
- Torres J., Mehandru S., Colombel J.F., Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741–1755. - PubMed
-
- Ordás I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W.J. Ulcerative colitis. Lancet. 2012;380:1606–1619. - PubMed
-
- Suzuki K., Murano T., Shimizu H., Ito G., Nakata T., Fujii S., Ishibashi F., Kawamoto A., Anzai S., Kuno R., Kuwabara K., Takahashi J., Hama M., Nagata S., Hiraguri Y., Takenaka K., Yui S., Tsuchiya K., Nakamura T., Ohtsuka K., Watanabe M., Okamoto R. Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol. 2018;53:1035–1047. - PMC - PubMed
-
- Dotti I., Mora-Buch R., Ferrer-Picón E., Planell N., Jung P., Masamunt M.C., Leal R.F., Martín de Carpi J., Llach J., Ordás I., Batlle E., Panés J., Salas A. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. - "VSports注册入口" PMC - PubMed
Publication types
VSports app下载 - MeSH terms
- "VSports注册入口" Actions
- "V体育安卓版" Actions
- "VSports在线直播" Actions
Substances
- "V体育2025版" Actions
- VSports注册入口 - Actions
"V体育2025版" LinkOut - more resources
Full Text Sources
"V体育平台登录" Medical
"VSports在线直播" Miscellaneous

