Single-cell transcriptome of early hematopoiesis guides arterial endothelial-enhanced functional T cell generation from human PSCs
- PMID: 34516916
- PMCID: PMC8442917
- DOI: 10.1126/sciadv.abi9787
Single-cell transcriptome of early hematopoiesis guides arterial endothelial-enhanced functional T cell generation from human PSCs
Abstract
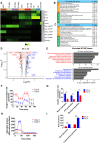
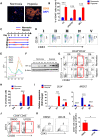
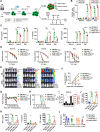
Hematopoietic differentiation of human pluripotent stem cells (hPSCs) requires orchestration of dynamic cell and gene regulatory networks but often generates blood cells that lack natural function. Here, we performed extensive single-cell transcriptomic analyses to map fate choices and gene expression patterns during hematopoietic differentiation of hPSCs and showed that oxidative metabolism was dysregulated during in vitro directed differentiation. Applying hypoxic conditions at the stage of endothelial-to-hematopoietic transition in vitro effectively promoted the development of arterial specification programs that governed the generation of hematopoietic progenitor cells (HPCs) with functional T cell potential. Following engineered expression of the anti-CD19 chimeric antigen receptor, the T cells generated from arterial endothelium-primed HPCs inhibited tumor growth both in vitro and in vivo VSports手机版. Collectively, our study provides benchmark datasets as a resource to further understand the origins of human hematopoiesis and represents an advance in guiding in vitro generation of functional T cells for clinical applications. .
Figures (VSports手机版)



References
-
- Hansen M., von Lindern M., van den Akker E., Varga E., Human-induced pluripotent stem cell-derived blood products: State of the art and future directions. FEBS Lett. 593, 3288–3303 (2019). - PubMed
-
- Sugimura R., Jha D. K., Han A., Soria-Valles C., da Rocha E. L., Lu Y. F., Goettel J. A., Serrao E., Rowe R. G., Malleshaiah M., Wong I., Sousa P., Zhu T. N., Ditadi A., Keller G., Engelman A. N., Snapper S. B., Doulatov S., Daley G. Q., Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017). - PMC - PubMed
-
- Gordon-Keylock S., Sobiesiak M., Rybtsov S., Moore K., Medvinsky A., Mouse extraembryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood 122, 2338–2345 (2013). - "VSports" PMC - PubMed
-
- Lancrin C., Mazan M., Stefanska M., Patel R., Lichtinger M., Costa G., Vargel O., Wilson N. K., Moroy T., Bonifer C., Gottgens B., Kouskoff V., Lacaud G., GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120, 314–322 (2012). - PubMed
Grants and funding (V体育安卓版)
LinkOut - more resources
V体育ios版 - Full Text Sources
Other Literature Sources
Molecular Biology Databases

