Farnesoid X receptor (FXR): Structures and ligands
- PMID: 33995909
- PMCID: PMC8091178
- DOI: 10.1016/j.csbj.2021.04.029
Farnesoid X receptor (FXR): Structures and ligands
Erratum in
-
Corrigendum to: "Farnesoid X receptor (FXR): Structures and ligands" [Comput. Struct. Biotechnol. J. 19 (2021) 2148-2159].Comput Struct Biotechnol J. 2022 Mar 1;20:1227-1228. doi: 10.1016/j.csbj.2022.02.029. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35317235 Free PMC article.
Abstract
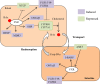
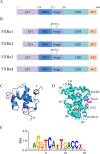
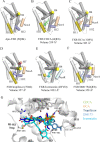
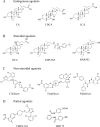
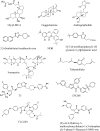
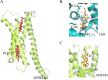
Farnesoid X receptor (FXR) is a bile acid activated nuclear receptor (BAR) and is mainly expressed in the liver and intestine. Upon ligand binding, FXR regulates key genes involved in the metabolic process of bile acid synthesis, transport and reabsorption and is also involved in the metabolism of carbohydrates and lipids. Because of its important functions, FXR is considered as a promising drug target for the therapy of bile acid-related liver diseases. With the approval of obeticholic acid (OCA) as the first small molecule to target FXR, many other small molecules are being evaluated in clinical trials VSports手机版. This review summarizes the structures of FXR, especially its ligand binding domain, and the development of small molecules (including agonists and antagonists) targeting FXR. .
Keywords: Agonists; Antagonists; Farnesoid X receptor; Ligand binding domain; Structure V体育安卓版. .
© 2021 The Author(s).
VSports在线直播 - Figures

References
-
- Lee F.Y., Lee H., Hubbert M.L., Edwards P.A., Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31(10):572–580. - PubMed
-
- Zhang Z.D., Cayting P., Weinstock G., Gerstein M. Analysis of nuclear receptor pseudogenes in vertebrates: how the silent tell their stories. Mol Biol Evol. 2008;25(1):131–143. - PubMed
-
- Zhang Y., Kast-Woelbern H.R., Edwards P.A. Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. J Biol Chem. 2003;278(1):104–110. - PubMed
Publication types
- "V体育安卓版" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
