Histone chaperone CAF-1 promotes HIV-1 latency by leading the formation of phase-separated suppressive nuclear bodies
- PMID: 33739466
- PMCID: PMC8126954 (V体育官网)
- DOI: 10.15252/embj.2020106632 (VSports在线直播)
"VSports最新版本" Histone chaperone CAF-1 promotes HIV-1 latency by leading the formation of phase-separated suppressive nuclear bodies
Abstract
HIV-1 latency is a major obstacle to achieving a functional cure for AIDS VSports手机版. Reactivation of HIV-1-infected cells followed by their elimination via immune surveillance is one proposed strategy for eradicating the viral reservoir. However, current latency-reversing agents (LRAs) show high toxicity and low efficiency, and new targets are needed to develop more promising LRAs. Here, we found that the histone chaperone CAF-1 (chromatin assembly factor 1) is enriched on the HIV-1 long terminal repeat (LTR) and forms nuclear bodies with liquid-liquid phase separation (LLPS) properties. CAF-1 recruits epigenetic modifiers and histone chaperones to the nuclear bodies to establish and maintain HIV-1 latency in different latency models and primary CD4+ T cells. Three disordered regions of the CHAF1A subunit are important for phase-separated CAF-1 nuclear body formation and play a key role in maintaining HIV-1 latency. Disruption of phase-separated CAF-1 bodies could be a potential strategy to reactivate latent HIV-1. .
Keywords: CAF-1; HIV-1 latency; epigenetic regulation; nuclear body; phase separation V体育安卓版. .
© 2021 The Authors.
Conflict of interest statement (V体育安卓版)
The authors declare that they have no conflict of interest.
Figures
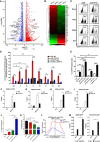
- A
RNA‐Seq of naïve and TNFα‐stimulated J‐Lat 10. 6 cells. Differentially expressed genes were filtered with log2FC of 1 and P value FDR cutoff of 0. 05. Upregulated and downregulated genes were labeled as red and blue dots, respectively. Representative genes were labeled aside corresponding dots V体育平台登录.
- B
RNA‐Seq result as in (A). Significantly changed genes were sorted out and plotted as heatmap. SEC16A indicated unchanged gene VSports注册入口.
- C
The GFP‐positive percentages of monoclonal sgCHAF1A and sgNT J‐Lat 10. 6 cell lines were shown in the top right corner of each flow cytometry figure. TNFα, SAHA, and JQ‐1 were used as supplements V体育官网入口.
- D
ChIP assay with antibody against CHAF1A was performed in J‐Lat 10. 6 cells VSports在线直播. All the ChIP‐qPCR DNA signals were normalized to siNC IgG of G5′. G5′ represented cellular DNA and viral 5′LTR junction; A: Nucleosome 0 assembly site; B: Nucleosome free region; C: Nucleosome 1 assembly site; V5: Viral 5′LTR and gag leader sequence junction; E represented envelope; V3: Viral poly purine tract and 3′LTR junction; G3′ represented viral 3′LTR and cellular DNA junction.
- E
The statistical graph of the result in (C).
- F–I
ChIP assays with antibodies against H3K9me3, H4K20me3, H3K4me3, and H3K9Acetyl were performed as in (D) V体育2025版. Only “C” position signals were showed and normalized to Input.
- J, K
J‐Lat 8.4 cells were treated with shCHAF1A lentiviruses and 5‐aza‐dC. The GFP‐positive cells and HIV‐1 LTR methyl‐CpGs percentages of different groups were plotted in (J) and (K), respectively.
- L
ATAC‐Seq was performed in WT and CHAF1A‐KO J‐Lat 10.6 cells. The relative tag densities of the pseudotyped HIV‐1 5′LTR integration site in each group were calculated. The highest tag density was set as 100. Figure showed 2 kb range centered the 5′LTR integration site. Dashed line represented HIV‐1 pseudotyped virus 5′LTR integration site.
- M
ChIP assays with antibodies against CDK9 and Pho‐Pol II were performed in siNC and siCHAF1A J‐Lat 10.6 cells. ChIP signals in each group were normalized to input.

- A
Mass spectrometry (MS) result of naïve and TNFα‐stimulated J‐Lat 10.6 cells. Significantly changed genes were plotted as heatmap. Representative genes were shown in table aside the heatmap. The heatmap scale represented fold change of gene expression (Min: −20 fold; Max: 20 fold).
- B, C
The GFP‐positive percentages of heterogeneous sgCHAF1A and sgNT J‐Lat 10.6 cell lines were shown as flow cytometry figure and statistical histogram. TNFα, SAHA and JQ‐1 were used as supplements.
- D
Fifteen LRAs targeting eight signaling pathways were used in heterogeneous and homogeneous CHAF1A‐KO J‐Lat 10.6 cell lines.
- E, F
Two pseudotyped HIV‐1 latency cell lines (J‐Lat‐NIB and J‐Lat‐NPB) were treated with sgCHAF1A lentiviruses, SAHA and JQ‐1. The GFP‐positive percentages of each group were measured. The backbones of each pseudotyped HIV‐1 were also shown.
- G–J
J‐Lat 6.3, 8.4, 9.2 and 15.4 were treated as in Fig 1C. The reactivation efficiencies of each group were indicated by the percentages of GFP‐positive cells.
- A
ChIP assay with antibody against CHAF1A was performed in TZM‐bl cells as in Fig 1D. ChIP‐qPCR DNA signals were normalized to Input. G5: cellular DNA and viral 5’LTR junction; L: luciferase region; G3: viral 3′LTR and cellular DNA junction. The other regions were as in Fig 1D.
- B–E
ChIP assays with antibodies against H3K9me, H3K9me2, H3K27me3 and H3K36me2 were performed as in Fig 1F.
- F
The schematic of CpGs on HIV‐1 LTR of J‐Lat 8.4. Eleven CpGs were shown in circles.
- G, H
The flow cytometry figures and Methyl‐CpGs graphs of J‐Lat 8.4 which were treated with shCHAF1A and 5‐aza‐dC, corresponding to statistical histograms in Fig 1J and K.
- I
RNA‐Seq result of wild‐type and CHAF1A‐KO J‐Lat 10.6. Differentially expressed genes were filtered with log2FC of 1 and PvalueFDR cutoff of 0.05. Upregulated and downregulated genes were labeled as red and blue dots, respectively. Representative genes were labeled aside corresponding dots.
- J
Flag‐tagged TRIM28, SUMO1, SUMO2 and SUMO4 were co‐overexpressed with HA‐tagged CHAF1A. HA‐tagged CHAF1A was IP with anti‐HA beads. Both total and IP samples were IB with anti‐HA, anti‐Flag and anti‐Actin antibodies.
- K
HA‐tagged CHAF1A and HA‐tagged GFP were co‐overexpressed with Flag‐tagged CDK9. HA‐tagged proteins were immunoprecipitated (IP) with anti‐HA beads. Both total and IP samples were immunoblotted (IB) with anti‐HA, anti‐Flag and anti‐GAPDH antibodies.
- L
HA‐tagged CDK9 was co‐overexpressed with Flag‐tagged SUMO4, UBC9, and TRIM28. siRNAs targeting TRIM28 and CHAF1A treated CDK9‐overexpressed HeLa cells, respectively. CDK9 was IP with anti‐HA beads. Both total and IP samples were IB with anti‐HA, anti‐Flag and anti‐GAPDH antibodies.
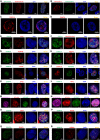
- A
GFP‐tagged CHAF1A was imaged with AF594‐tagged antibody against CHAF1A.
- B
RFP‐tagged CHAF1A was imaged with AF488‐tagged antibody against CHAF1A.
- C, D
The endogenous CHAF1A was imaged before (C) and after (D) CHAF1A knockdown in J‐Lat 10.6 cells.
- E, F
Coilin and Pho‐Ser5‐RNAP II were immunostained with corresponding antibodies and imaged with CHAF1A, respectively.
- G–L
CHAF1B, RBBP4, PCNA, SUV39H2, HP1β, and HP1γ was co‐overexpressed with CHAF1A, respectively. SIM images were captured for each combination.
- M
RFP‐tagged PCNA and GFP‐tagged DNMT1 were imaged with AF647‐tagged antibody against CHAF1A simultaneously. DAPI was used to dye DNA.
- N
RFP‐tagged HP1α, GFP‐tagged DNMT1, and BFP‐tagged PCNA were imaged with AF647‐tagged antibody against CHAF1A simultaneously.
- O–R
CBX4, SUZ12, SUMO1, and SUMO2 were imaged with CHAF1A, respectively.

- A
The distribution of CHAF1A‐PIP2‐C. PIP2: the second PCNA‐interacting protein motif which located after KER. C: CHAF1A C‐terminal.
- B
The co‐localization of RFP‐tagged SUMO1, SUMO2 and SUMO4 with GFP‐tagged CHAF1A‐dDS13.
- C
The distribution of CHAF1A‐dDS13‐dSIM.
- D–F
The enrichment of CHAF1A, H4K20me3 and H3K9Acetyl on HIV‐1 LTR in different CHAF1A status.
 "VSports注册入口"
"VSports注册入口"
- A
The procedure of CHAF1A‐mediated HIV‐1 integration restriction. On Day −4, primary CD4+ T cells were isolated from PBMCs of healthy donors, followed by the activation of PHA. On Day −2, endogenous CHAF1A was knocked down by shCHAF1A lentiviruses. Two days later, shluc‐ and shCHAF1A‐treated cells were proceeded to the infection of pseudotyped HIV‐1. Another 4 days later, the percentages of GFP‐positive cells, which indicated the reactivation efficiency, were measured by FCM.
- B
The schematic of pseudotyped HIV‐1 backbone used in (A).
- C, D
The FCM figures and statistical histogram of shluc‐ and shCHAF1A‐treated groups.
- E
The procedure of CHAF1A‐mediated delay of HIV‐1 entering into latency. On Day −4, primary CD4+ T cells were isolated and activated by PHA. On Day −2, activated CD4+ T cells were washed and infected with pseudotyped HIV‐1. Another 2 days later, cells were infected with shluc or shCHAF1A lentiviruses. In the following 2 weeks, the percentages of GFP‐positive cells were measured by FCM every 4 days.
- F
The statistical histogram of the results in (E). Data showed results from three different healthy donors.
- G
The procedure of CHAF1A‐mediated HIV‐1 latency in wild‐type HIV‐1 latency model. The procedure was similar as in Fig 7A, except that the virus used in (G) was wild‐type HIV‐1 and the infected cells were treated with 5 μM AZT during the 2 weeks of resting status.
- H
The schematic of GFP‐conjugated wild‐type HIV‐1 backbone used in (G).
- I, J
The representative FCM figures and statistical scatter plot of the results in (G).
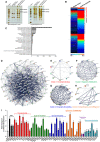
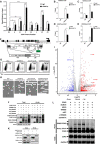
- A
HA‐tagged CHAF1A and CHAF1B were co‐overexpressed in HEK293T cells. Forty‐eight hours post‐transfection, cells were lysed and immunoprecipitated (IP) with anti‐HA beads. The control groups were transfected with empty vector. The IP samples were proceeded to SDS–PAGE and developed with silver staining. Data showed both short and long development results. The whole lane of each group was cut into several gel slices and proceeded to in‐gel digestion and LC‐MS/MS.
- B
The heatmap of CHAF1A‐ and CHAF1B‐enriched proteins. Gray bar‐covered genes represented nucleus proteins. Pink bar‐covered genes represented chromatin binding proteins.
- C
Gene Ontology (GO) analysis of significantly enriched proteins utilizing PANTHER classification system.
- D
STRING network analysis of enriched proteins with the interaction confidence of 0.7.
- E–H
k‐means clustering of significantly enriched proteins. Four major clusters were classified: chromatin binding, epigenetic modification, chromatin remodeling, ubiquitination, and SUMOylation.
- I
siRNAs targeting each CAF‐1 subunits and corresponding enriched genes were transfected into TZM‐bl cell lines, respectively. The fold change of luciferase expression of each groups was normalized to siNC.

- A
Super‐resolution SIM image of CHAF1A in HEK293T cells. DAPI was used to dye DNA which was colored into blue. FITC‐tagged antibody was used to label endogenous CHAF1A which was colored into green.
- B
Super‐resolution cSTORM image of CHAF1A in HEK293T cells. Two degrees of amplification were performed to show CAF‐1 bodies.
- C, D
ImmunoFISH images of CAF‐1 bodies and HIV‐1 genomic DNA within J‐Lat 10.6. Naïve cells were treated with DMSO. Activated cells were acquired by treating with TNFα.
- E
RFP‐tagged CHAF1A and GFP‐tagged PML body component SP100 were co‐overexpressed in HEK293T cells and imaged by SIM.
- F
GFP‐tagged CHAF1A and RFP‐tagged BRD4 were co‐overexpressed in HEK293T cells and imaged by SIM.
- G–M
SUV39H1, HP1α, DNMT1, MBD1, RING1B, EZH2, and SUMO4 were co‐overexpressed with CHAF1A in HEK293T cells, respectively. SIM imaging was performed for each combination.
- N–P
GFP‐tagged CHAF1A was overexpressed in HEK293T cells. CF568‐tagged antibodies against H3K9me3, H3K27me3, and H3K4me3 were used to treat IF samples, respectively. CHAF1A and each histone modification were imaged by SIM.

- A
FRAP images of CAF‐1 bodies to indicate the internal diffusion property. GFP‐tagged CHAF1A was overexpressed in HEK293T cells. Twenty‐four hours later, live cells were proceeded to time series imaging. One of the CAF‐1 bodies was bleached with intense laser pulse. Images of live cell were captured every 4 s. One of the unbleached CAF‐1 bodies was used as control. Data represented eight time points. At least three cells of each sample were treated with intense laser pulse to obtain bleached CAF‐1 bodies.
- B
Representative images of two CAF‐1 bodies fusing into one.
- C
Representative images of one CAF‐1 body splitting into two.
- D
Live cells were imaged every 2 s in time series. 1,6‐hexanediol was added into the cell incubation well carefully at time point of 5th second. Images were captured for 3 min.
- E
GFP‐tagged CHAF1A‐N‐NLS and CHAF1A‐C were imaged with RFP‐tagged CHAF1A, respectively. N: CHAF1A N‐terminal. NLS: nuclear localization signal of CHAF1A, which was presented within C‐terminal. C: CHAF1A C‐terminal.
- F
Upper panel showed WT‐CHAF1A bodies. Lower panel showed dLLPS‐CHAF1A which was CHAF1A‐Q34A‐I116A‐K395D‐R402D‐K409D.
- G–I
GFP‐tagged CHAF1A‐IDR and CHAF1A‐mIDR were purified in vitro. Ten micromole of each protein was incubated with gradient droplet formation buffer. The droplets of CHAF1A‐IDR appeared in NaCl concentration of 125 mM or lower (G). No CHAF1A‐mIDR droplet was detected within any NaCl concentration (I). The volumes of CHAF1A‐IDR droplets increased with the increase of protein concentration (H). Left subpanels within each panel represented images of protein droplets in different buffers. Right subpanels within each panel represented statistical analysis results of droplets areas and numbers.
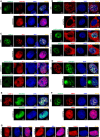
- A
The distribution of CHAF1B in the presence of CHAF1A (upper panel) and in the absence of endogenous CHAF1A (lower panel).
- B
First panel: the co‐localization of PCNA and CHAF1A. Second panel: the distribution of PCNA in the absence of endogenous CHAF1A. Third panel: the distribution of PCNA in the presence of CHAF1A‐dPIP1 mutant and the absence of endogenous CHAF1A. Fourth panel: the co‐localization of PCNA and overexpressed DNMT1 in the absence of endogenous CHAF1A.
- C
Upper two panels: the distribution of HP1α in the presence or absence of CHAF1A. Lower two panels: the distribution of HP1α in the presence of CHAF1A‐dHP1BD or CHAF1A‐DS2‐dHP1BD. Both images of lower two panels were in the absence of endogenous CHAF1A.
- D–F
The distribution of SUV39H1 (D), SUV39H2 (E) and MBD1 (F) in the presence or absence of CHAF1A.
- G
The distribution of SUMO paralogs in the absence of CHAF1A.
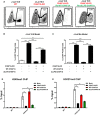
- A
Flow cytometry figures of CAF‐1 body rescue experiment. First figure: Wild‐type J‐Lat 10.6. Second figure: CHAF1A knockout J‐Lat 10.6. Third figure: CHAF1A‐KO J‐Lat 10.6 was re‐introduced with wild‐type CHAF1A. Fourth figure: CHAF1A‐KO J‐Lat 10.6 was re‐introduced with dLLPS‐CHAF1A which harbored five mutations. The percentages of GFP‐positive cells were labeled in the top right corner of each figure.
- B
The statistical histogram of the results in (A).
- C
Rescue experiment in heterogeneous Jurkat latency model which were performed as in (A).
- D, E
The enrichment of H3K9me3 and H3K27me3 on HIV‐1 LTR in different CHAF1A status. The reactivation efficiency in each group was indicated by the percentage of GFP‐positive cells.
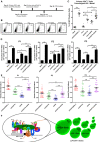
The procedure of CHAF1A depletion‐mediated HIV‐1 reactivation in primary CD4+ T cells. On Day −4, primary CD4+ T cells were isolated from PBMCs of healthy donors, and activated by PHA. On Day 0, activated CD4+ T cells were washed and infected with pseudotyped HIV‐1. Another 2 days later, cells were treated with shluc and shCHAF1A lentiviruses, respectively. Two weeks later, one part of naïve CD4+ T cells was treated with αCD3/αCD28/IL‐2. One part of shluc‐treated cells and one part of shCHAF1A‐treated cells were further treated with 500 nM SAHA. On day 16, all the cells were proceeded to FCM analysis.
The flow cytometry figures of each group. The reactivation efficiency was indicated by the GFP‐positive cells percentages which were labeled on the top right corner.
The statistical scatter plot of the results in (B).
The amounts of intracellular HIV‐1 RNAs of latently infected primary CD4+ T cells which were isolated from PBMCs of HIV‐1‐infected individuals. αCD3/αCD28/IL‐2 treatment was performed as positive control. shCHAF1A lentiviruses were packaged to knock down endogenous CHAF1A. shluc lentiviruses treatment was performed as negative control. SAHA was added as LRA supplement.
Envelope V1–V3 region of HIV‐1 RNAs in (D) was reverse‐transcribed and PCR‐amplified. The PCR products were proceeded to TA‐cloning. At least 60 single clones were picked from each group and sequenced. These sequences and the standard HIV‐1 sequence HXB2 were aligned. Genetic diversity index of each sequence was calculated for each group.
The schematic of CAF‐1 body‐mediated HIV‐1 latency. Detailed information for the schematic was indicated in discussion.
References
-
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 - VSports - PMC - PubMed
-
- Blazkova J, Trejbalova K, Gondois‐Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J et al (2009) CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5: e1000554 - PMC (V体育官网入口) - PubMed
-
- Boehm D, Jeng M, Camus G, Gramatica A, Schwarzer R, Johnson JR, Hull PA, Montano M, Sakane N, Pagans S et al (2017) SMYD2‐mediated histone methylation contributes to HIV‐1 latency. Cell Host Microbe 21: 569–579.e566 - V体育ios版 - PMC - PubMed
V体育官网 - Publication types
MeSH terms
- V体育ios版 - Actions
- "V体育平台登录" Actions
- Actions (V体育安卓版)
- Actions (V体育官网入口)
Substances
- VSports注册入口 - Actions
Associated data
- VSports最新版本 - Actions
V体育官网入口 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials (VSports app下载)

