Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization
- PMID: 33526676
- PMCID: PMC8017965
- DOI: 10.1073/pnas.2017709118
V体育官网入口 - Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization
Abstract
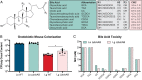
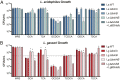
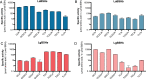
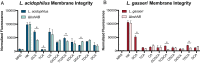
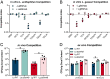
Primary bile acids (BAs) are a collection of host-synthesized metabolites that shape physiology and metabolism. BAs transit the gastrointestinal tract and are subjected to a variety of chemical transformations encoded by indigenous bacteria. The resulting microbiota-derived BA pool is a mediator of host-microbiota interactions VSports手机版. Bacterial bile salt hydrolases (BSHs) cleave the conjugated glycine or taurine from BAs, an essential upstream step for the production of deconjugated and secondary BAs. Probiotic lactobacilli harbor a considerable number and diversity of BSHs; however, their contribution to Lactobacillus fitness and colonization remains poorly understood. Here, we define and compare the functions of multiple BSHs encoded by Lactobacillus acidophilus and Lactobacillus gasseri Our genetic and biochemical characterization of lactobacilli BSHs lend to a model of Lactobacillus adaptation to the gut. These findings deviate from previous notions that BSHs generally promote colonization and detoxify bile. Rather, we show that BSH enzymatic preferences and the intrinsic chemical features of various BAs determine the toxicity of these molecules during Lactobacillus growth. BSHs were able to alter the Lactobacillus transcriptome in a BA-dependent manner. Finally, BSHs were able to dictate differences in bacterial competition in vitro and in vivo, defining their impact on BSH-encoding bacteria within the greater gastrointestinal tract ecosystem. This work emphasizes the importance of considering the enzymatic preferences of BSHs alongside the conjugated/deconjugated BA-bacterial interaction. These results deepen our understanding of the BA-microbiome axis and provide a framework to engineer lactobacilli with improved bile resistance and use probiotics as BA-altering therapeutics. .
Keywords: Acidophilus; Lactobacillus; bile acid; bile salt hydrolase; gasseri V体育安卓版. .
Copyright © 2021 the Author(s) V体育ios版. Published by PNAS. .
V体育ios版 - Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Li J.et (V体育官网) al. .; MetaHIT Consortium , An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014). - PubMed
-
- Pasolli E., et al. ., Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019). - PMC (VSports最新版本) - PubMed
-
- Cummings J. H., Macfarlane G. T., Role of intestinal bacteria in nutrient metabolism. JPEN J. Parenter. Enteral Nutr. 21, 357–365 (1997). - PubMed
-
- Wilson I. D., Nicholson J. K., Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222 (2017). - PMC (VSports) - PubMed
-
- Jacobs J. P., et al. ., A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2, 750–766 (2016). - "VSports" PMC - PubMed
Publication types (VSports app下载)
"V体育ios版" MeSH terms
- V体育官网入口 - Actions
- Actions (V体育安卓版)
- Actions (V体育官网)
- "V体育官网入口" Actions
"V体育官网入口" Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

