"VSports手机版" Cellular Heterogeneity-Adjusted cLonal Methylation (CHALM) improves prediction of gene expression
- PMID: 33452255
- PMCID: PMC7811027 (V体育平台登录)
- DOI: "VSports手机版" 10.1038/s41467-020-20492-7
Cellular Heterogeneity-Adjusted cLonal Methylation (CHALM) improves prediction of gene expression
Abstract
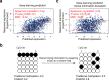
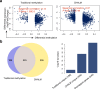
Promoter DNA methylation is a well-established mechanism of transcription repression, though its global correlation with gene expression is weak VSports手机版. This weak correlation can be attributed to the failure of current methylation quantification methods to consider the heterogeneity among sequenced bulk cells. Here, we introduce Cell Heterogeneity-Adjusted cLonal Methylation (CHALM) as a methylation quantification method. CHALM improves understanding of the functional consequences of DNA methylation, including its correlations with gene expression and H3K4me3. When applied to different methylation datasets, the CHALM method enables detection of differentially methylated genes that exhibit distinct biological functions supporting underlying mechanisms. .
Conflict of interest statement (V体育ios版)
After completing the current studies at Baylor College of Medicine, J. X. became a full-time employee at Helio Health. W V体育安卓版. L. is a consultant for Helio Health and ChosenMed. The remaining authors declare no competing interests.
Figures



References
-
- Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. - VSports手机版 - DOI - PubMed
Publication types
- "VSports在线直播" Actions
MeSH terms
- V体育官网入口 - Actions
- VSports - Actions
- VSports在线直播 - Actions
- VSports注册入口 - Actions
Substances
- VSports - Actions
Grants and funding
LinkOut - more resources
"V体育2025版" Full Text Sources
Other Literature Sources
Molecular Biology Databases

