Characterization and generation of human definitive multipotent hematopoietic stem/progenitor cells
- PMID: 33298886
- PMCID: PMC7705709
- DOI: 10.1038/s41421-020-00213-6
"VSports最新版本" Characterization and generation of human definitive multipotent hematopoietic stem/progenitor cells
VSports手机版 - Abstract
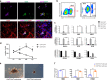
Definitive hematopoiesis generates hematopoietic stem/progenitor cells (HSPCs) that give rise to all mature blood and immune cells, but remains poorly defined in human. Here, we resolve human hematopoietic populations at the earliest hematopoiesis stage by single-cell RNA-seq. We characterize the distinct molecular profiling between early primitive and definitive hematopoiesis in both human embryonic stem cell (hESC) differentiation and early embryonic development VSports手机版. We identify CD44 to specifically discriminate definitive hematopoiesis and generate definitive HSPCs from hESCs. The multipotency of hESCs-derived HSPCs for various blood and immune cells is validated by single-cell clonal assay. Strikingly, these hESCs-derived HSPCs give rise to blood and lymphoid lineages in vivo. Lastly, we characterize gene-expression dynamics in definitive and primitive hematopoiesis and reveal an unreported role of ROCK-inhibition in enhancing human definitive hematopoiesis. Our study provides a prospect for understanding human early hematopoiesis and a firm basis for generating blood and immune cells for clinical purposes. .
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
VSports - Other Literature Sources
Molecular Biology Databases
Miscellaneous

