lncRNA SNHG10 Promotes the Proliferation and Invasion of Osteosarcoma via Wnt/β-Catenin Signaling
- PMID: 33251045
- PMCID: "V体育2025版" PMC7674123
- DOI: 10.1016/j.omtn.2020.10.010
lncRNA SNHG10 Promotes the Proliferation and Invasion of Osteosarcoma via Wnt/β-Catenin Signaling (VSports在线直播)
Abstract
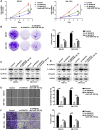
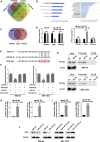
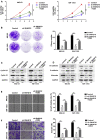
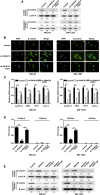
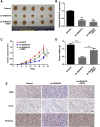
Uncontrolled growth and an enforced epithelial-mesenchymal transition (EMT) process contribute to the poor survival rate of patients with osteosarcoma (OS). Long noncoding RNAs (lncRNAs) have been reported to be involved in the development of OS. However, the significant role of lncRNA SNHG1O on regulating proliferation and the EMT process of OS cells remains unclear. In this study, quantitative real-time PCR and fluorescence in situ hybridization (FISH) results suggested that SNHG10 levels were significantly increased in OS compared with healthy tissues. In vitro experiments (including colony formation, CCK-8, wound healing, and transwell assays) and in vivo experiments indicated that downregulation of SNHG10 significantly suppressed the proliferation and invasion of OS cells. Luciferase reporter assay and RNA immunoprecipitation (RIP) assay confirmed that SNHG10 could regulate FZD3 levels through sponging microRNA 182-5p (miR-182-5p). In addition, the SNHG10/miR-182-5p/FZD3 axis could further promote the β-catenin transfer into nuclear accumulation to maintain the activation of the Wnt singling pathway. Together, our results established that SNHG10 has an important role in promoting OS growth and invasion VSports手机版. By sponging miR-182-5p, SNHG10 can increase FZD3 expression and further maintain the activation of Wnt/β-catenin singling pathway in OS cells. .
Keywords: EMT; SNHG10; Wnt/β-catenin; lncRNA; osteosarcoma V体育安卓版. .
© 2020 The Authors.
Figures (V体育平台登录)
V体育ios版 - References
-
- Tian W., Li Y., Zhang J., Li J., Gao J. Combined analysis of DNA methylation and gene expression profiles of osteosarcoma identified several prognosis signatures. Gene. 2018;650:7–14. - PubMed
-
- Berner K., Johannesen T.B., Berner A., Haugland H.K., Bjerkehagen B., Bøhler P.J., Bruland O.S. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54:25–33. - PMC (VSports最新版本) - PubMed
-
- Yu D., Kahen E., Cubitt C.L., McGuire J., Kreahling J., Lee J., Altiok S., Lynch C.C., Sullivan D.M., Reed D.R. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci. Rep. 2015;5:16991. - "VSports最新版本" PMC - PubMed
-
- Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. - PubMed
LinkOut - more resources
VSports注册入口 - Full Text Sources
"V体育ios版" Miscellaneous

