phyloFlash: Rapid Small-Subunit rRNA Profiling and Targeted Assembly from Metagenomes (V体育平台登录)
- PMID: 33109753
- PMCID: PMC7593591 (VSports在线直播)
- DOI: 10.1128/mSystems.00920-20
phyloFlash: Rapid Small-Subunit rRNA Profiling and Targeted Assembly from Metagenomes
"VSports" Abstract
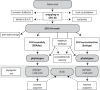
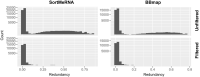
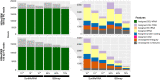
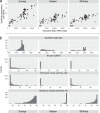
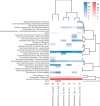
The small-subunit rRNA (SSU rRNA) gene is the key marker in molecular ecology for all domains of life, but it is largely absent from metagenome-assembled genomes that often are the only resource available for environmental microbes. Here, we present phyloFlash, a pipeline to overcome this gap with rapid, SSU rRNA-centered taxonomic classification, targeted assembly, and graph-based binning of full metagenomic assemblies. We show that a cleanup of artifacts is pivotal even with a curated reference database. With such a filtered database, the general-purpose mapper BBmap extracts SSU rRNA reads five times faster than the rRNA-specialized tool SortMeRNA with similar sensitivity and higher selectivity on simulated metagenomes. Reference-based targeted assemblers yielded either highly fragmented assemblies or high levels of chimerism, so we employ the general-purpose genomic assembler SPAdes. Our optimized implementation is independent of reference database composition and has satisfactory levels of chimera formation. phyloFlash quickly processes Illumina (meta)genomic data, is straightforward to use, even as part of high-throughput quality control, and has user-friendly output reports. The software is available at https://github. com/HRGV/phyloFlash (GPL3 license) and is documented with an online manual. IMPORTANCE To track organisms across all domains of life, the SSU rRNA gene is the gold standard. Many environmental microbes are known only from high-throughput sequence data, but the SSU rRNA gene, the key to visualization by molecular probes and link to existing literature, is often missing from metagenome-assembled genomes (MAGs). The easy-to-use phyloFlash software suite tackles this gap with rapid, SSU rRNA-centered taxonomic classification, targeted assembly, and graph-based linking to MAGs VSports手机版. Starting from a cleaned reference database, phyloFlash profiles the taxonomic diversity and assembles the sorted SSU rRNA reads. The phyloFlash design is domain agnostic and covers eukaryotes, archaea, and bacteria alike. phyloFlash also provides utilities to visualize multisample comparisons and to integrate the recovered SSU rRNAs in a metagenomics workflow by linking them to MAGs using assembly graph parsing. .
Keywords: SSU; gene assembly; metagenomics; taxonomic profiling. V体育安卓版.
Copyright © 2020 Gruber-Vodicka et al.
"VSports在线直播" Figures

References (VSports)
-
- Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542. doi:10.1038/s41564-017-0012-7. - VSports最新版本 - DOI - PubMed
"V体育官网入口" LinkOut - more resources
Full Text Sources (VSports手机版)
V体育平台登录 - Miscellaneous

