PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis
- PMID: 32929201
- PMCID: V体育平台登录 - PMC7653546
- DOI: 10.1038/s41556-020-0575-z
PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis
Erratum in
-
Author Correction: PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis.Nat Cell Biol. 2020 Nov;22(11):1396. doi: 10.1038/s41556-020-00599-1. Nat Cell Biol. 2020. PMID: 33033376
Abstract
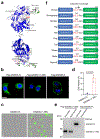
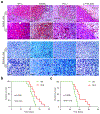
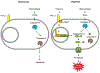
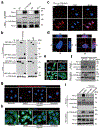
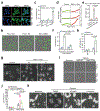
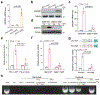
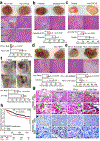
Although pyroptosis is critical for macrophages against pathogen infection, its role and mechanism in cancer cells remains unclear. PD-L1 has been detected in the nucleus, with unknown function. Here we show that PD-L1 switches TNFα-induced apoptosis to pyroptosis in cancer cells, resulting in tumour necrosis. Under hypoxia, p-Stat3 physically interacts with PD-L1 and facilitates its nuclear translocation, enhancing the transcription of the gasdermin C (GSDMC) gene. GSDMC is specifically cleaved by caspase-8 with TNFα treatment, generating a GSDMC N-terminal domain that forms pores on the cell membrane and induces pyroptosis VSports手机版. Nuclear PD-L1, caspase-8 and GSDMC are required for macrophage-derived TNFα-induced tumour necrosis in vivo. Moreover, high expression of GSDMC correlates with poor survival. Antibiotic chemotherapy drugs induce pyroptosis in breast cancer. These findings identify a non-immune checkpoint function of PD-L1 and provide an unexpected concept that GSDMC/caspase-8 mediates a non-canonical pyroptosis pathway in cancer cells, causing tumour necrosis. .
Conflict of interest statement
Declaration of Interests
The authors have no conflicts of interest to declare.
Figures







"V体育ios版" Comment in
-
PD-L1 controls cancer pyroptosis.Nat Cell Biol. 2020 Oct;22(10):1157-1159. doi: 10.1038/s41556-020-00582-w. Nat Cell Biol. 2020. PMID: 32943765 No abstract available.
References
-
- Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). - PubMed (V体育2025版)
-
- Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). - PubMed
-
- Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). - PubMed (VSports在线直播)
-
- Ding J et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016). - PubMed
Publication types
- "V体育ios版" Actions
- "V体育ios版" Actions
MeSH terms
- "VSports手机版" Actions
- "V体育ios版" Actions
- Actions (VSports最新版本)
- V体育2025版 - Actions
- "VSports手机版" Actions
- V体育ios版 - Actions
- VSports手机版 - Actions
- Actions (V体育官网入口)
- "V体育2025版" Actions
- VSports最新版本 - Actions
"V体育安卓版" Substances
- V体育ios版 - Actions
"VSports" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育官网入口)
Medical
Molecular Biology Databases
"VSports最新版本" Research Materials
"VSports app下载" Miscellaneous

